SPI1 Antibody
-
货号:CSB-PA003887
-
规格:¥880
-
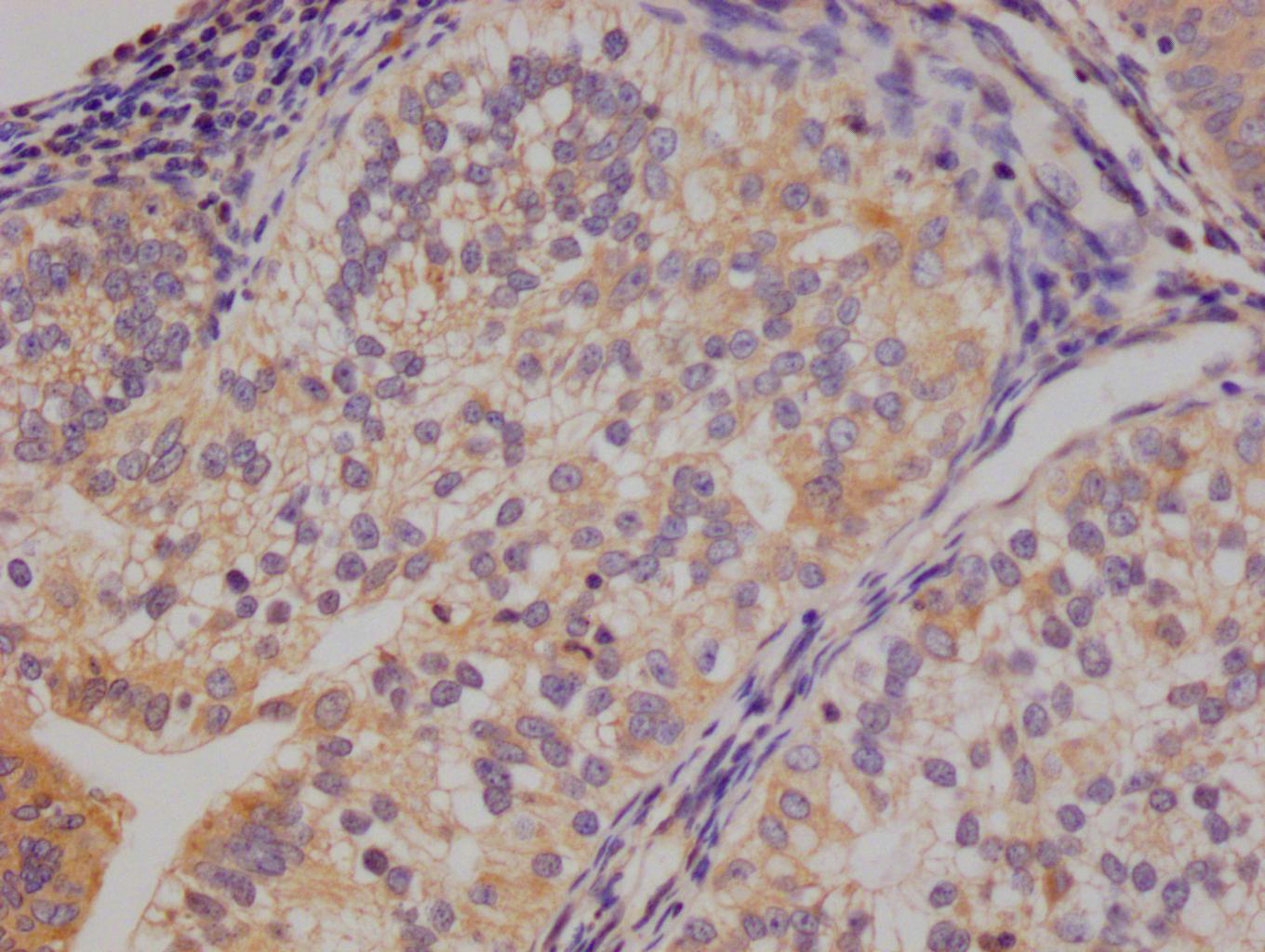
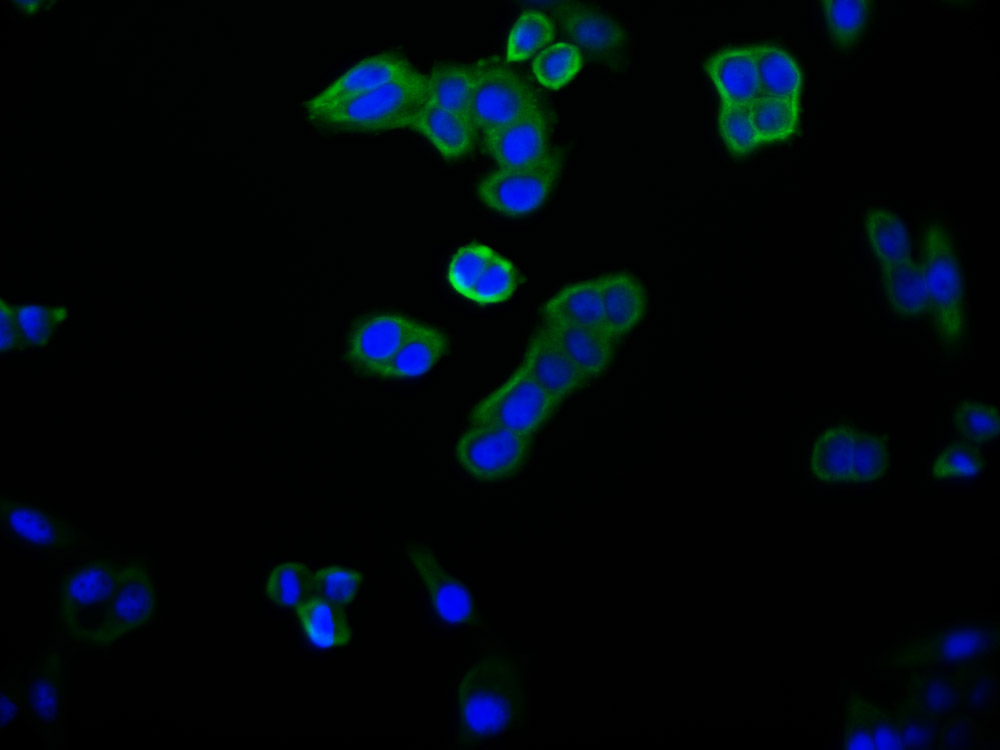
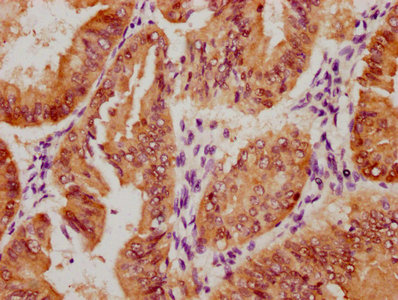
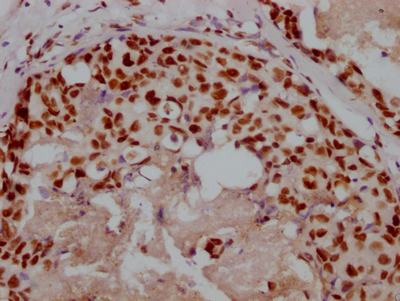
图片:
-
其他:
产品详情
-
Uniprot No.:P17947
-
基因名:
-
别名:transcription factor spi1 antibody; 31 kDa Transforming Protein antibody; 31 kDa-transforming protein antibody; cb1086 antibody; Hematopoietic transcription factor PU.1 antibody; OF antibody; oncogene spi1 antibody; PU.1 antibody; SFFV virus-induced murine erythroleukemia oncogene, mouse, homolog of antibody; SFPI1 antibody; si:by184l24.2 antibody; SPI 1 antibody; SPI 1 proto oncogene antibody; SPI A antibody; Spi1 antibody; SPI1_HUMAN antibody; Spleen focus forming virus (SFFV) proviral integration oncogene spi1 antibody; Spleen focus forming virus proviral integration oncogene spi1 antibody; Transcription factor PU.1 antibody
-
宿主:Rabbit
-
反应种属:Human,Mouse,Rat,Monkey
-
免疫原:Synthesized peptide derived from the Internal region of Human PU.1.
-
免疫原种属:Homo sapiens (Human)
-
标记方式:Non-conjugated
-
抗体亚型:IgG
-
纯化方式:The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using epitope-specific immunogen.
-
浓度:It differs from different batches. Please contact us to confirm it.
-
保存缓冲液:Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide.
-
产品提供形式:Liquid
-
应用范围:WB, IHC, IF, ELISA
-
推荐稀释比:
Application Recommended Dilution WB 1:500-1:2000 IHC 1:100-1:300 IF 1:200-1:1000 ELISA 1:40000 -
Protocols:
-
储存条件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
货期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
相关产品
靶点详情
-
功能:Binds to the PU-box, a purine-rich DNA sequence (5'-GAGGAA-3') that can act as a lymphoid-specific enhancer. This protein is a transcriptional activator that may be specifically involved in the differentiation or activation of macrophages or B-cells. Also binds RNA and may modulate pre-mRNA splicing.
-
基因功能参考文献:
- these results suggest that attenuating PU.1 may be a valid therapeutic approach to limit microglial-mediated inflammatory responses in Alzheimer's disease PMID: 30124174
- PU.1 3'UTR attenuates TNFalphainduced proliferation and cytokine release of RAFLS by acting as a ceRNA for FOXO3 to regulate miR155 activity. PMID: 29693176
- Inhibition of endogenous miR-155 in B cells of rheumatoid arthritis patients restores PU.1 and reduces production of antibodies. PMID: 27671860
- PU.1 binds to OX40L promoter in dendritic cells. PMID: 27708417
- These results bring indirect evidence that leukemia develops from cells which have bypassed Spi1-induced senescence. Overall, our results reveal senescence as a Spi1-induced anti-proliferative mechanism that may be a safeguard against the development of acute myeloid leukemia. PMID: 28912174
- In contrast, expression of Spi1/PU.1 in a Fli1 producing erythroleukemia cell line in which fli1 is activated, resulted in increased proliferation through activation of growth promoting proteins MAPK, AKT, cMYC and JAK2 PMID: 28586009
- Data show that protein phosphatase-1 alpha (PP1alpha) is required to maintain checkpoint kinase 1 (CHK1) in a dephosphorylated state and for the accelerated replication fork progression in Spi1/PU.1 transcription factor-overexpressing cells. PMID: 28415748
- the results indicate that PU.1 may be a critical factor for the innate defense against A. fumigatus, and may therefore be a potential target for the prophylaxis and treatment of IPA. PMID: 28440496
- PU.1 supports TRAIL-induced cell death by inhibiting RelA-mediated cell survival and inducing DR5 expression. PMID: 28362429
- PU.1 directly activates the expression of HOTAIRM1 through binding to the regulatory region of HOTAIRM1 during granulocytic differentiation. PMID: 27146823
- PU.1 and IL-9 may play a role in AD pathogenesis and relate to disease severity and clinical eruption types. PMID: 28229452
- PU.1 has a role in tumor suppression in PEL and its down-regulation is associated with PEL development. PMID: 28481873
- PU.1-induced apoptosis in myeloma cells is associated with IRF4 downregulation and subsequent IRF7 upregulation. PMID: 28368411
- Most cases of histiocytic sarcoma expressed histiocytic markers CD68 (6 of 7 cases), CD163 (5 of 5 cases), and PU.1 (3 of 4 cases). PMID: 28805986
- findings highlight a unique role of SPI1 fusions in high-risk pediatric T cell acute lymphoblastic leukemia PMID: 28671687
- Alzheimer's disease heritability was enriched within the PU.1 cistrome, implicating a myeloid PU.1 target gene network in AD. PMID: 28628103
- expression of an essential mediator of neutrophil terminal differentiation, the ets transcription factor PU.1, was significantly decreased in Hbb(th3/+) neutrophils in beta-thalassemia PMID: 28325862
- RUNX1 overexpression induced partial DNA demethylation at SPI1 proximal promoter. PMID: 28376714
- This study demonstrated the novel role of PU.1 in the immune response to A. fumigatus through upregulation of Dectin-1 expression and its translocation to the nucleus in A. fumigatus-stimulated THP-1 cells. PMID: 27306059
- PU.1 is an important modulator of VDR signaling in monocytes. PMID: 28232093
- Forced FOG1 protein expression in K562 erythroleukemia cells induced the expression of SLC4A1 protein, but repressed that of transcription factor PU.1. PMID: 28216155
- Moreover, the expression of a cell proliferation marker Ki67 was significantly decreased in tumors from the mice not taking doxycycline, compared with that of tumors from the mice continuously taking doxycycline. The present data strongly suggest that PU.1 functions as a tumor suppressor of myeloma cells in vivo. PMID: 28347818
- we demonstrated that miR-22 promoted monocyte/macrophage differentiation, and MECOM (EVI1) mRNA is a direct target of miR-22 and MECOM (EVI1) functions as a negative regulator in the differentiation.The miR-22-mediated MECOM degradation increased c-Jun but decreased GATA2 expression, which results in increased interaction between c-Jun and PU.1 PMID: 27617961
- we conclude that PU.1 transactivates the pIII through direct binding to Ets-motifs in the promoter in pDCs PMID: 27105023
- Our results suggest the existence of a Vav1/PU.1/miR-142-3p network that supports all-trans retinoic acid -induced differentiation in acute promyelocytic leukemia -derived cells PMID: 27480083
- Our data suggest that E2A antagonism of PU.1 activity contributes to its ability to commit multipotential hematopoietic progenitors to the lymphoid lineages. PMID: 26942192
- This study showed that HCV infection might abrogate NK cytotoxic potential through altering PU.1, NKG2D receptor and perforin molecules. PMID: 26429314
- SPI1-GFI1B transcriptional network is an important regulatory axis in acute myeloid leukemia as well as in the development of erythroid versus myelomonocytic cell fate PMID: 26851695
- The GATA-1-mediated inhibition of PU.1 gene transcription in human AML-erythroleukemias mediated through the URE represents important mechanism that contributes to PU.1 downregulation and leukemogenesis that is sensitive to DNA demethylation therapy PMID: 27010793
- PU.1 downregulation was noted in B-CLL/SLL samples positive for the adverse prognostic markers CD38 and ZAP-70. PMID: 26261072
- PU.1 recruitment coupled with increased histone acetylation induces gene expression and activates a monocyte/macrophage transcriptional programme. PMID: 26126967
- This study demonstrated positive regulation of monocyte/macrophage differentiation by lnc-MC and uncovered an elaborate regulation mechanism composed of PU.1, lnc-MC, miR-199a-5p, and ACVR1B. PMID: 26149389
- Collectively, IMiDs exert demethylation activity through inhibiting DNMT1, 3a, and 3b, and up-regulating PU.1 expression, which may be one of the mechanisms of the anti-myeloma activity of IMiDs. PMID: 26657848
- Loss of PU.1 expression is associated with Hepatocellular Carcinoma. PMID: 25987019
- PU.1 suppressive target gene, metallothionein 1G, inhibits retinoic acid-induced NB4 cell differentiation. PMID: 25072246
- This review summarizes current knowledge and ideas of molecular mechanisms by which PU.1 controls hematopoiesis and suppresses leukemia. [review] PMID: 25205721
- A novel network has been described in acute myeloid leukemia in which FLT3-ITD signaling induces oncogenic miR-155 by p65 and STAT5 thereby targeting transcription factor PU.1. PMID: 25092144
- The increased CITED2 expression in acute myeloid leukemia results in better hematopoietic stem cell survival, lower PU.1 levels, and perturbed myeloid differentiation program that contributes to leukemia persistence. PMID: 25184385
- Runx-dependent PU.1 chromatin interaction and transcription of PU.1 are essential for both normal and leukemia stem cells. PMID: 25185713
- PU.1- targeted genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. PMID: 24028770
- DNA complex may be relevant to an emerging role of PU.1, but not Ets-1, as a pioneer transcription factor in vivo PMID: 24952944
- Data show that CCCTC-binding factor (CTCF) together with ISWI ATPase SMARCA5 and members of the Cohesin complex associate with the SPI1 protein is disrupted in acute myeloid leukemia (AML) blasts. PMID: 24498324
- The PU.1-regulated MAP1S gene is implicated in neutrophil differentiation and autophagy control. PMID: 25043887
- hnRNP K and PU.1 act synergistically during granulocytic differentiation, hnRNP K seems to have a negative effect on PU.1 activity during monocytic maturation PMID: 25005557
- Data indicate that transcription factors RUNX1 and PU.1 cooperated to exchange corepressors for coactivators, and deficiency of RUNX1, frequent in leukemia, caused aberrant recruitment of specific corepressors instead of coactivators to PU.1. PMID: 24695740
- IL-32theta; reduces PKCdelta-mediated phosphorylation of PU.1, resulting in attenuation of IL-1beta production PMID: 24996056
- HSF1 appears as a fine-tuning regulator of SPI1/PU.1 expression at the transcriptional and post-translational levels during macrophage differentiation of monocytes. PMID: 24504023
- our findings demonstrate that PU.1 contributes to the development of MLL leukemia, partially via crosstalk with the MEIS/HOX pathway. PMID: 24445817
- Given the importance of C/EBPs and PU.1 in myeloid development, these results, thus, suggest that restoration of the normal function of the myeloid cell transcriptional machinery is a major molecular mechanism underlying the differentiation induction PMID: 24379003
- Mice with PU.1 deficiency in T cells were protected from colitis, whereas treatment with antibody to IL-9 suppressed colitis PMID: 24908389
显示更多
收起更多
-
亚细胞定位:Nucleus.
-
蛋白家族:ETS family
-
数据库链接:
HGNC: 11241
OMIM: 165170
KEGG: hsa:6688
STRING: 9606.ENSP00000227163
UniGene: Hs.502511
Most popular with customers
-
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IF, FC
Species Reactivity: Human, Mouse, Rat
-
Phospho-YAP1 (S127) Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC
Species Reactivity: Human
-
-
-
-
-