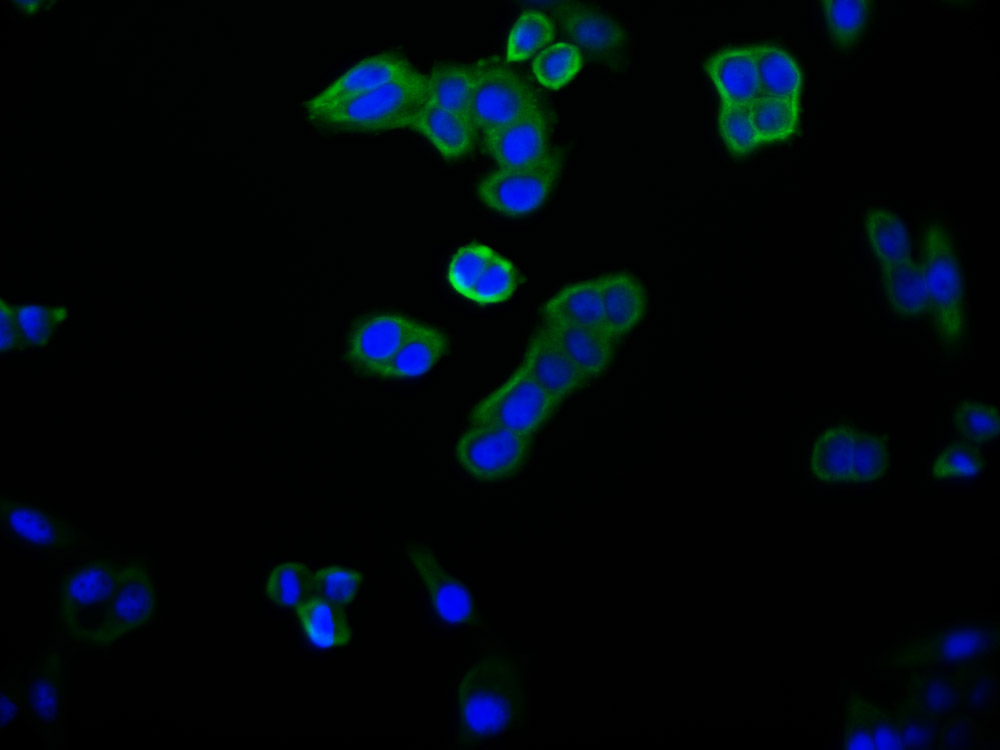
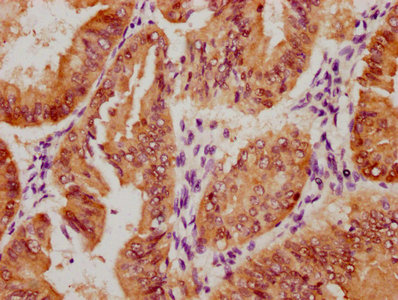
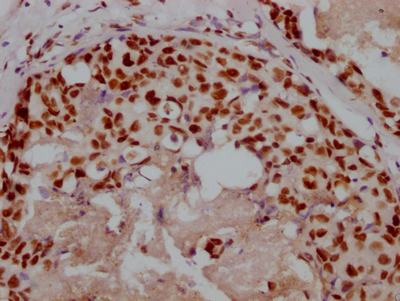
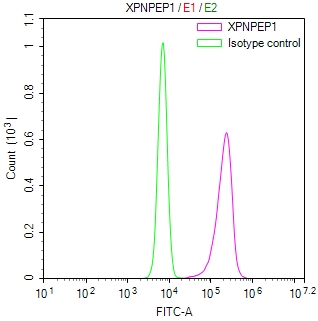
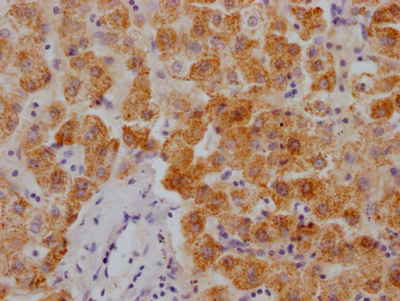
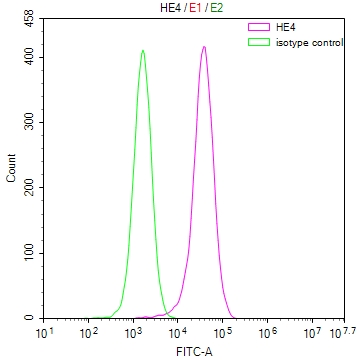
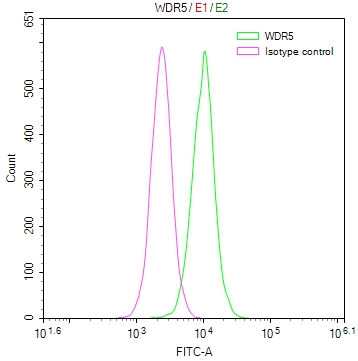
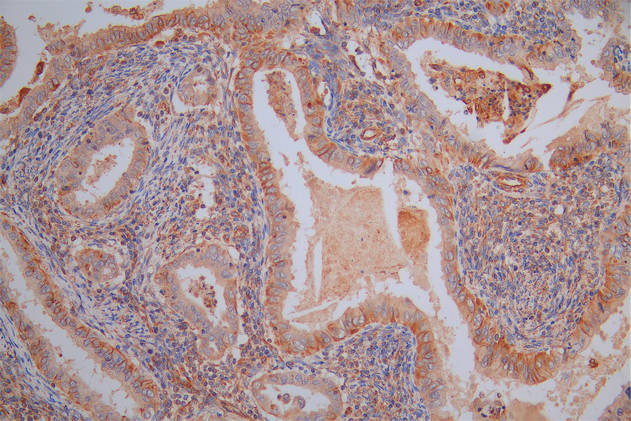
DDB1 Antibody
-
货号:CSB-PA006581GA01HU
-
规格:¥3,900
-
其他:
产品详情
-
Uniprot No.:Q16531
-
基因名:
-
别名:Damage specific DNA binding protein 1 antibody; Damage-specific DNA-binding protein 1 antibody; DDB 1 antibody; DDB p127 subunit antibody; Ddb1 antibody; DDB1_HUMAN antibody; DDBa antibody; DNA damage binding protein 1 antibody; DNA damage-binding protein 1 antibody; DNA damage-binding protein a antibody; HBV X-associated protein 1 antibody; UV damaged DNA binding factor antibody; UV damaged DNA binding protein 1 antibody; UV DDB 1 antibody; UV DDB1 antibody; UV-damaged DNA-binding factor antibody; UV-damaged DNA-binding protein 1 antibody; UV-DDB 1 antibody; X associated protein 1 antibody; XAP 1 antibody; XAP-1 antibody; XAP1 antibody; Xeroderma pigmentosum group E complementing protein antibody; Xeroderma pigmentosum group E-complementing protein antibody; XPCe antibody; XPE antibody; XPE BF antibody; XPE binding factor antibody; XPE-BF antibody; XPE-binding factor antibody
-
宿主:Rabbit
-
反应种属:Human,Mouse,Rat
-
免疫原:Human DDB1
-
免疫原种属:Homo sapiens (Human)
-
抗体亚型:IgG
-
纯化方式:Antigen Affinity Purified
-
浓度:It differs from different batches. Please contact us to confirm it.
-
保存缓冲液:PBS with 0.1% Sodium Azide, 50% Glycerol, pH 7.3. -20°C, Avoid freeze / thaw cycles.
-
产品提供形式:Liquid
-
应用范围:ELISA,WB,IHC
-
Protocols:
-
储存条件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
货期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
相关产品
靶点详情
-
功能:Protein, which is both involved in DNA repair and protein ubiquitination, as part of the UV-DDB complex and DCX (DDB1-CUL4-X-box) complexes, respectively. Core component of the UV-DDB complex (UV-damaged DNA-binding protein complex), a complex that recognizes UV-induced DNA damage and recruit proteins of the nucleotide excision repair pathway (the NER pathway) to initiate DNA repair. The UV-DDB complex preferentially binds to cyclobutane pyrimidine dimers (CPD), 6-4 photoproducts (6-4 PP), apurinic sites and short mismatches. Also functions as a component of numerous distinct DCX (DDB1-CUL4-X-box) E3 ubiquitin-protein ligase complexes which mediate the ubiquitination and subsequent proteasomal degradation of target proteins. The functional specificity of the DCX E3 ubiquitin-protein ligase complex is determined by the variable substrate recognition component recruited by DDB1. DCX(DDB2) (also known as DDB1-CUL4-ROC1, CUL4-DDB-ROC1 and CUL4-DDB-RBX1) may ubiquitinate histone H2A, histone H3 and histone H4 at sites of UV-induced DNA damage. The ubiquitination of histones may facilitate their removal from the nucleosome and promote subsequent DNA repair. DCX(DDB2) also ubiquitinates XPC, which may enhance DNA-binding by XPC and promote NER. DCX(DTL) plays a role in PCNA-dependent polyubiquitination of CDT1 and MDM2-dependent ubiquitination of TP53 in response to radiation-induced DNA damage and during DNA replication. DCX(ERCC8) (the CSA complex) plays a role in transcription-coupled repair (TCR). The DDB1-CUL4A-DTL E3 ligase complex regulates the circadian clock function by mediating the ubiquitination and degradation of CRY1. DDB1-mediated CRY1 degradation promotes FOXO1 protein stability and FOXO1-mediated gluconeogenesis in the liver.
-
基因功能参考文献:
- These results suggest that different DDB1-CUL4 associated factors play distinct roles in human lung adenocarcinoma development. PMID: 28336923
- The DDB1 is acetylated and acetylation promotes DDB1 binding to CUL4. PMID: 28886238
- Results revealed a function independent of its transcriptional activity, as TTF-1 was found to interact with DDB1 and block its binding to CHK1, which in turn attenuated ubiquitylation and subsequent degradation of CHK1. PMID: 28192407
- SIRT7 inhibits TR4 degradation by deacetylation of DDB1. PMID: 28623141
- the c-Abl non-receptor kinase phosphorylates DDB1 at residue Tyr-316 to recruit a small regulatory protein, DDA1, leading to increased substrate ubiquitination PMID: 28087699
- knockdown of DCAF7 reduced the degradation of DNA ligase I in response to inhibition of proliferation and replacement of ubiquitylated lysine residues reduced the in vitro ubiquitylation of DNA ligase I by Cul4-DDB1 and DCAF7. In contrast, a different E3 ubiquitin ligase regulates FEN-1 turnover. PMID: 27573245
- This study presents the crystal structure of the DDB1-DCAF1-HIV-1-Vpr-uracil-DNA glycosylase (cyclin U) complex. PMID: 27571178
- Our data are consistent with the idea that the CUL4A/B-DDB1-CRBN complex catalyses the polyubiquitination and thus controls the degradation of CLC-1 channels. PMID: 26021757
- These results revealed a novel role of DDB in H3K56Ac deacetylation during early step of NER and the existence of active functional cross-talk between DDB-mediated damage recognition and H3K56Ac deacetylation. PMID: 26255936
- The identification of Vpr mutants which associate with DCAF1 but only poorly with DDB1 suggests that DCAF1 is necessary but is not sufficient for the Vpr association with DDB1-containing E3 ligase complex. PMID: 24912982
- Data support a model wherein DDB1 and DDB2 cooperate to repress Bcl-2 transcription. DDB2 recognizes and binds to the Bcl-2 P1 promoter, and HDAC1 is recruited through the DDB1 subunit associated with DDB2 to deacetylate histone H3K9. PMID: 24249678
- The study presents the crystal structure of human CRBN bound to DDB1 and the drug lenalidomide. PMID: 25108355
- CUL4A-DDB1-Rbx1 E3 ligase controls the quality of the PTS2 receptor Pex7p. PMID: 24989250
- structures of the DDB1-CRBN complex bound to thalidomide, lenalidomide and pomalidomide PMID: 25043012
- In the three intrinsically IMiD-resistant cell lines that clearly express detectable levels of cereblon, the absence of CRBN and DDB1 mutations suggest that potential cereblon-independent mechanisms of resistance exist PMID: 24166296
- UV-DDB examines sites on DNA in discrete steps before forming long-lived, nonmotile UV-DDB dimers (DDB1-DDB2)2 at sites of damage. PMID: 24760829
- p73 interacts with the CDL4A complex by binding directly to DDB1. The CDL4A complex is able to monoubiquitylate p73, negatively affecting its transcriptional function. PMID: 23085759
- As a molecular adaptor, Vpr enhanced the interaction between TERT and the VPRBP substrate receptor of the DYRK2-associated EDD-DDB1-VPRBP E3 ligase complex, resulting in increased ubiquitination of TERT. PMID: 23612978
- Data indicate that Dyrk2 phosphorylates TERT protein, which is then associated with the EDD-DDB1-VprBP E3 ligase complex for subsequent ubiquitin-mediated TERT protein degradation. PMID: 23362280
- Our findings suggest that DDB1 is a cellular substrate of NS3/4A required for Hepatitis c viurs replication. PMID: 23137809
- The EZH2-DCAF1/DDB1/CUL4 represents a previously unrecognized methylation-dependent ubiquitination machinery specifically recognizing "methyl degron"; nonhistone protein stability can be dynamically regulated in a methylation-dependent manner. PMID: 23063525
- potential GRK5 interacting proteins and the association of GRK5 with DDB1 in cell and the regulation of GRK5 level by DDB1-CUL4 ubiquitin ligase complex-dependent proteolysis pathway PMID: 22952844
- Findings indicate structural and conformational insights of the DDB1-CUL4A(DDB2) E3 ligase, with significant implications for the regulation and overall organization of the proteins responsible for initiation of nucleotide-excision repair (NER) pathway. PMID: 22822215
- The data suggested that HomolD-containing promoters require the RNA polymerase II machinery and the proteins DDB1 and RECQL for accurate transcription. PMID: 22705827
- Hepatitis B virus regulatory HBx protein binding to DDB1 is required but is not sufficient for maximal HBV replication. PMID: 22342275
- crystals of CSA-DDB1 had unit-cell parameters a = b = 142.03, c = 250.19 A and diffracted to 2.9 A resolution on beamline ID14-1 PMID: 22232169
- Studies indicate the modular architecture of DDB1-CUL4 in complex with DDB2, CSA and CDT2 in DNA repair of UV-induced DNA lesions. PMID: 21550341
- damage-specific DNA binding protein 1 is essential to regulation of p27(kip1) turnover after a mild DNA damage PMID: 21237244
- the CUL4A.DDB1 E3 complex is important for regulation of RASSF1A during mitosis, and it may contribute to inactivation of RASSF1A and promoting cell cycle progression PMID: 21205828
- This review focuses on Vpr and its HIV2/SIV counterparts, Vpx and Vpr, which all engage the DDB1.Cullin4 ubiquitin ligase complex through the DCAF1 adaptor protein. PMID: 20347598
- DDB1 modulates the function of APC/C(Cdh1) in a manner independent of the Cul4-DDB1 complex PMID: 20395298
- The data suggest that DDB1 could potentially be developed into biomarkers of resistance to acyl sulfonamide-based cancer drugs. This will require clinical validation in a series of patients treated with R3200. PMID: 19723642
- Studies indicate that CUL4 uses a large beta-propeller protein, DDB1, as a linker to interact with a subset of WD40 proteins. PMID: 19818632
- Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation PMID: 12034848
- findings substantiate the physical and functional connection between the hepatitis B virus X protein and the DDB1-DDB2 heterodimer, leading to the regulation of the pool of the viral protein PMID: 12050362
- These findings indicate that hepatitis B virus X protein acts through a pathway that involves a DDB2-independent nuclear function of DDB1 and that this activity will depend on the relative concentration of DDB1 and DDB2 in cells. PMID: 12151405
- essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. DDB1 may form a multiprotein complex with STAT1, STAT2, and V for this degradation. PMID: 12388698
- SV5-V and HBx have evolved to bind DDB1 to achieve distinct functions in their life cycle, both by a mechanism that does not involve DDB2. PMID: 12743284
- DET1 promotes ubiquitination and degradation of c-Jun by assembling a multisubunit ubiquitin ligase containing DNA Damage Binding Protein-1 (DDB1), cullin 4A (CUL4A), Regulator of Cullins-1 (ROC1), and constitutively photomorphogenic-1 PMID: 14739464
- Damaged DNA binding protein 1 is a component of the centromere complex in interphase cells. PMID: 15009096
- results show that HBx in association with DDB1 acts in the nucleus and stimulates hepatitis B virus replication mainly by enhancing viral mRNA levels PMID: 15767425
- DDB1-DDB2 protein complex recognizes DNA mismatches and lesions PMID: 16223728
- PCNA is involved in mediating Cdt1 degradation by the Cul4-Ddb1 ligase in response to DNA damage. PMID: 16407242
- Cdt1 degradation requires predominant use of the PCNA/Cul4/Ddb1 ubiquitin ligase pathway after DNA damage PMID: 16407252
- Monoubiquitinated histone H2A in native chromatin coimmunoprecipitates with the endogenous DDB1-CUL4A(DDB2) complex in response to UV irradiation. PMID: 16473935
- The F-box protein Skp2, in addition to utilizing Cul1-Skp1, utilizes Cul4A-DDB1 to induce proteolysis of p27Kip1. PMID: 16537899
- This study uncovers CUL4-DDB-ROC1 as a histone ubiquitin ligase and demonstrate that histone H3 and H4 ubiquitylation participates in the cellular response to DNA damage. PMID: 16678110
- PCNA, L2DTL and the DDB1-CUL4A complex play critical and differential roles in regulating the protein stability of p53 and MDM2/HDM2 in unstressed and stressed cells. PMID: 16861890
- L2DTL and PCNA interact with CUL4/DDB1 complexes and are involved in CDT1 degradation after DNA damage. PMID: 16861906
- Results suggest that DDB1 prevents DNA lesions from accumulating in replicating human cells, in part by regulating Cdt1 degradation. PMID: 16940174
显示更多
收起更多
-
亚细胞定位:Cytoplasm. Nucleus.
-
蛋白家族:DDB1 family
-
数据库链接:
HGNC: 2717
OMIM: 600045
KEGG: hsa:1642
STRING: 9606.ENSP00000301764
UniGene: Hs.290758
Most popular with customers
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IF, FC
Species Reactivity: Human, Mouse, Rat
-
Phospho-YAP1 (S127) Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC
Species Reactivity: Human
-
-
-
-
-
-