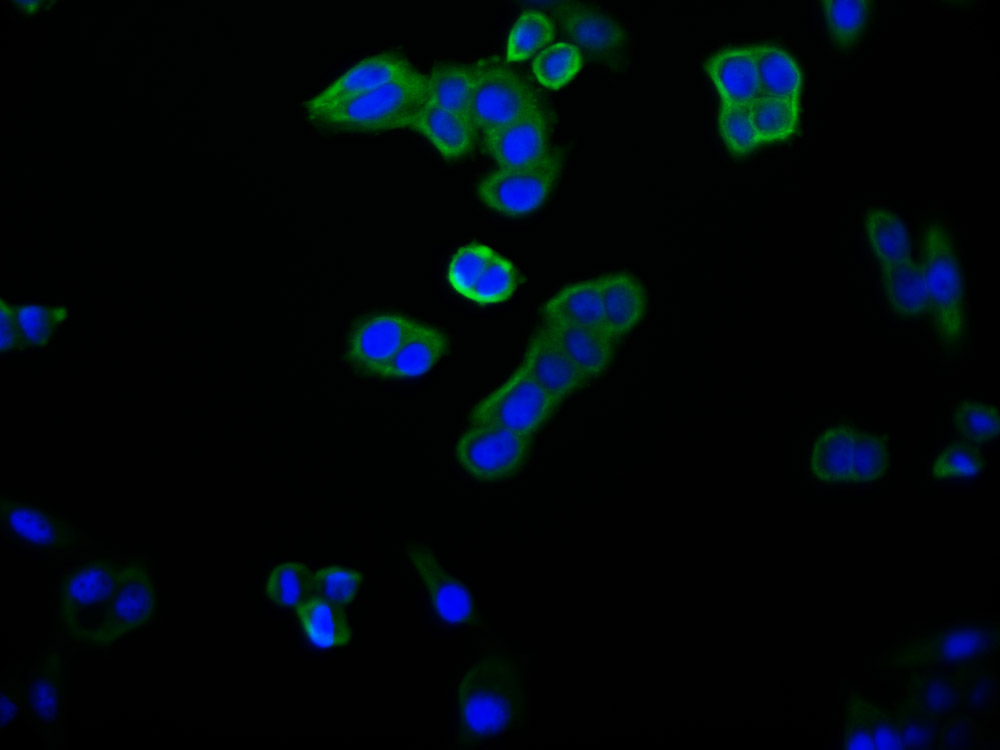
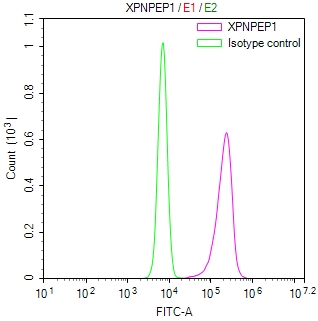
CRY2 Antibody
-
货号:CSB-PA673229ESR2HU
-
规格:¥440
-
促销:
-
图片:
-
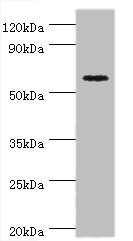
Western blot
All lanes: Cryptochrome-2 antibody at 14μg/ml + HepG2 whole cell lysate
Secondary
Goat polyclonal to rabbit IgG at 1/10000 dilution
Predicted band size: 67, 61 kDa
Observed band size: 67 kDa -
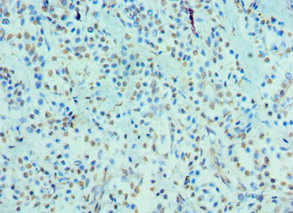
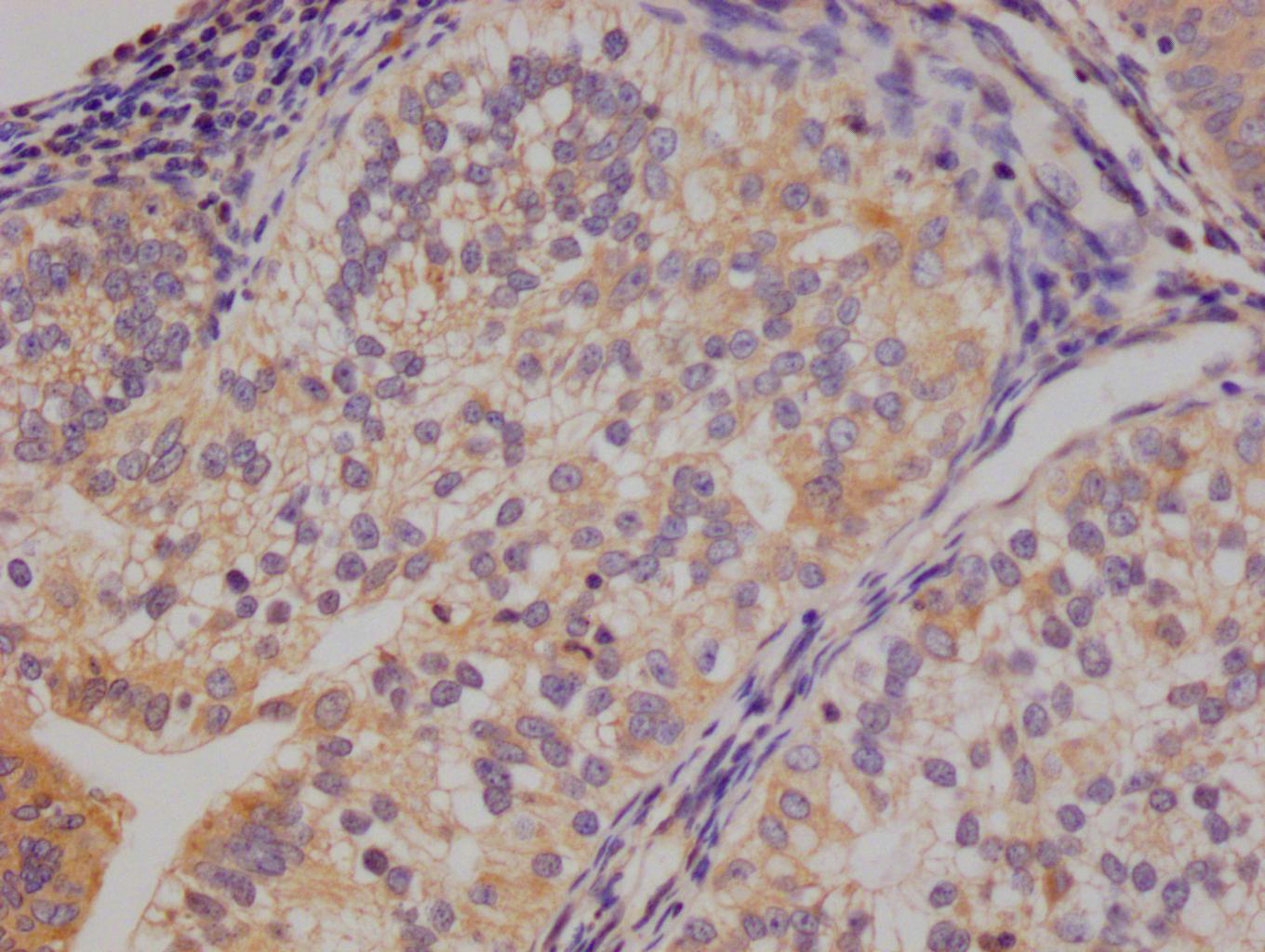
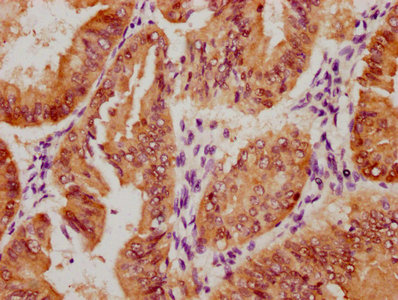
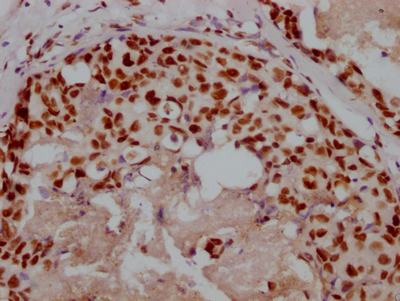
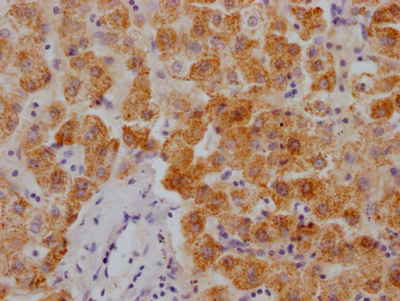
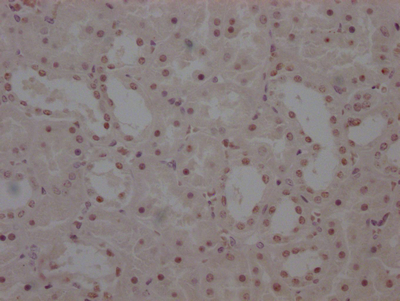
Immunohistochemistry of paraffin-embedded human breast cancer using CSB-PA673229ESR2HU at dilution of 1:100
-
-
其他:
产品详情
-
产品名称:Rabbit anti-Homo sapiens (Human) CRY2 Polyclonal antibody
-
Uniprot No.:Q49AN0
-
基因名:CRY2
-
别名:cry2 antibody; CRY2_HUMAN antibody; cryptochrome 2 (photolyase like) antibody; Cryptochrome 2 antibody; Cryptochrome-2 antibody; FLJ10332 antibody; growth inhibiting protein 37 antibody; HCRY2 antibody; KIAA0658 antibody; PHLL2 antibody; Photolyase like antibody
-
宿主:Rabbit
-
反应种属:Human
-
免疫原:Recombinant Human Cryptochrome-2 protein (434-593AA)
-
免疫原种属:Homo sapiens (Human)
-
标记方式:Non-conjugated
-
克隆类型:Polyclonal
-
抗体亚型:IgG
-
纯化方式:Antigen Affinity Purified
-
浓度:It differs from different batches. Please contact us to confirm it.
-
保存缓冲液:PBS with 0.02% sodium azide, 50% glycerol, pH7.3.
-
产品提供形式:Liquid
-
应用范围:ELISA, WB, IHC
-
推荐稀释比:
Application Recommended Dilution WB 1:200-1:1000 IHC 1:20-1:200 -
Protocols:
-
储存条件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
货期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
相关产品
靶点详情
-
功能:Transcriptional repressor which forms a core component of the circadian clock. The circadian clock, an internal time-keeping system, regulates various physiological processes through the generation of approximately 24 hour circadian rhythms in gene expression, which are translated into rhythms in metabolism and behavior. It is derived from the Latin roots 'circa' (about) and 'diem' (day) and acts as an important regulator of a wide array of physiological functions including metabolism, sleep, body temperature, blood pressure, endocrine, immune, cardiovascular, and renal function. Consists of two major components: the central clock, residing in the suprachiasmatic nucleus (SCN) of the brain, and the peripheral clocks that are present in nearly every tissue and organ system. Both the central and peripheral clocks can be reset by environmental cues, also known as Zeitgebers (German for 'timegivers'). The predominant Zeitgeber for the central clock is light, which is sensed by retina and signals directly to the SCN. The central clock entrains the peripheral clocks through neuronal and hormonal signals, body temperature and feeding-related cues, aligning all clocks with the external light/dark cycle. Circadian rhythms allow an organism to achieve temporal homeostasis with its environment at the molecular level by regulating gene expression to create a peak of protein expression once every 24 hours to control when a particular physiological process is most active with respect to the solar day. Transcription and translation of core clock components (CLOCK, NPAS2, ARNTL/BMAL1, ARNTL2/BMAL2, PER1, PER2, PER3, CRY1 and CRY2) plays a critical role in rhythm generation, whereas delays imposed by post-translational modifications (PTMs) are important for determining the period (tau) of the rhythms (tau refers to the period of a rhythm and is the length, in time, of one complete cycle). A diurnal rhythm is synchronized with the day/night cycle, while the ultradian and infradian rhythms have a period shorter and longer than 24 hours, respectively. Disruptions in the circadian rhythms contribute to the pathology of cardiovascular diseases, cancer, metabolic syndromes and aging. A transcription/translation feedback loop (TTFL) forms the core of the molecular circadian clock mechanism. Transcription factors, CLOCK or NPAS2 and ARNTL/BMAL1 or ARNTL2/BMAL2, form the positive limb of the feedback loop, act in the form of a heterodimer and activate the transcription of core clock genes and clock-controlled genes (involved in key metabolic processes), harboring E-box elements (5'-CACGTG-3') within their promoters. The core clock genes: PER1/2/3 and CRY1/2 which are transcriptional repressors form the negative limb of the feedback loop and interact with the CLOCK|NPAS2-ARNTL/BMAL1|ARNTL2/BMAL2 heterodimer inhibiting its activity and thereby negatively regulating their own expression. This heterodimer also activates nuclear receptors NR1D1/2 and RORA/B/G, which form a second feedback loop and which activate and repress ARNTL/BMAL1 transcription, respectively. CRY1 and CRY2 have redundant functions but also differential and selective contributions at least in defining the pace of the SCN circadian clock and its circadian transcriptional outputs. Less potent transcriptional repressor in cerebellum and liver than CRY1, though less effective in lengthening the period of the SCN oscillator. Seems to play a critical role in tuning SCN circadian period by opposing the action of CRY1. With CRY1, dispensable for circadian rhythm generation but necessary for the development of intercellular networks for rhythm synchrony. May mediate circadian regulation of cAMP signaling and gluconeogenesis by blocking glucagon-mediated increases in intracellular cAMP concentrations and in CREB1 phosphorylation. Besides its role in the maintenance of the circadian clock, is also involved in the regulation of other processes. Plays a key role in glucose and lipid metabolism modulation, in part, through the transcriptional regulation of genes involved in these pathways, such as LEP or ACSL4. Represses glucocorticoid receptor NR3C1/GR-induced transcriptional activity by binding to glucocorticoid response elements (GREs). Represses the CLOCK-ARNTL/BMAL1 induced transcription of BHLHE40/DEC1. Represses the CLOCK-ARNTL/BMAL1 induced transcription of NAMPT. Represses PPARD and its target genes in the skeletal muscle and limits exercise capacity. Represses the transcriptional activity of NR1I2.
-
基因功能参考文献:
- CRY2 levels (ng/mL) were markedly higher in both Metabolic syndrome (MetS) groups (non-diabetic and pre-diabetic/diabetic) (all with p-value < 0.001). A reciprocal melatonin-CRY2 relationship was observed in the MetS (non-diabetic) group (p-value = 0.003). PMID: 29813030
- CRY2 may be an anti-oncogene in osteosarcoma. PMID: 29879092
- data indicate an oncogenic role of miR-181d in CRC by promoting glycolysis, and miR-181d/CRY2/FBXL3/c-myc feedback loop might be a therapeutic target for patients with CRC. PMID: 28749470
- In the longitudinal analysis, CRY2 SNP rs61884508 was protective from worsening of problematicity of seasonal variations of mood disorder. In the cross-sectional analysis, CRY2 SNP rs72902437 showed evidence of association with problematicity of seasonal variations, as did SNP rs1554338 (in the MAPK8IP1 and downstream of CRY2). PMID: 27267441
- The earlier reported association of CRY2 variants with dysthymia was confirmed and extended to major depressive disorder. PMID: 27721187
- These results demonstrate that CRY2 stability controlled by FBXL3 plays a key role in the regulation of human sleep wake behavior. PMID: 27529127
- The FOXM1 is a negative regulator of CRY2 in breast cancer via enhancing methylation in CRY2 promoter and its high expression is an independent predictor of favorable MR-free survival in ER+ breast cancer patients. PMID: 28579430
- CRY2 and FBXL3 cooperatively degrade c-MYC preventing the development of cancer. PMID: 27840026
- For the first time, we show that Cry 2 rs2292910 and MTNR1B rs3781638 are associated with osteoporosis in a Chinese geriatric cohort. PMID: 26564225
- data may point to CRY2 as a novel switch in hepatic fuel metabolism promoting triglyceride storage and, concomitantly, limiting glucose production PMID: 26726810
- Data indicate that cryptochrome 2 (CRY2) knockdown leads to chemosensitivity of colorectal cancer cell lines. PMID: 25855785
- CRY2 and REV-ERB ALPHA as the clock genes upregulated in obesity during the 24 h period and that REV-ERB ALPHA is an important gene associated with MS. PMID: 25365257
- These findings suggest that the core circadian gene CRY2 is associated with breast cancer progression and prognosis, and that knockdown of CRY2 causes the epigenetic dysregulation of genes involved in cancer-relevant pathways PMID: 25740058
- CRY2 genetic variants associate with the depressive episodes in a range of mood disorder; CRY2 is hypothesized to a key to the resetting of clocks throughout and play a leading role in the antidepressant effect of total sleep deprivation PMID: 24099037
- Four CRY2 genetic variants (rs10838524, rs7121611, rs7945565, rs1401419) associated significantly with dysthymia (false discovery rate q<0.05). PMID: 23951166
- Our data indicate that variants in the circadian-related genes CRY2 and MTNR1B may affect long-term changes in energy expenditure, and dietary fat intake may modify the genetic effects. PMID: 24335056
- These data document for the first time that the expression of BMAL1, PER3, PPARD and CRY2 genes is altered in gestational diabetes compared to normal pregnant women. deranged expression of clock genes may play a pathogenic role in GDM. PMID: 23323702
- Reduced cry2 expression is associated with glioma. PMID: 23317246
- Genetic variants of CRY2 associate with seasonal affective disorder or winter depression. PMID: 22538398
- the associations of GLIS3-rs7034200 and CRY2-rs11605924 with fasting glucose, beta cell function, and type 2 diabetes PMID: 21747906
- Human CRY2 has the molecular capability to function as a light-sensitive magnetosensor. PMID: 21694704
- Lean individuals exhibited significant (P < 0.05) temporal changes of core clock (PER1, PER2, PER3, CRY2, BMAL1, and DBP) and metabolic (REVERBalpha, RIP140, and PGC1alpha) genes. PMID: 21411511
- CRY2 gene variation was associated with bipolar disorder. PMID: 20856823
- Data suggest that CRY2 locus is associated with vulnerability for depression. PMID: 20195522
- findings suggest a role for CRY2 in breast tumorigenesis PMID: 20233903
- Data suggest a limited, but potentially important role for CRY2 in the regulation of DNA damage repair and the maintenance of genomic stability in breast cancer. PMID: 20334671
- CRY2 binds to DNA and in particular to single-stranded DNA with moderately high affinity and to DNA containing (6-4) photoproducts with higher affinity, but it has no DNA repair activity. PMID: 12627958
- Transcripts of hCry2 underwent circadian oscillation. The expression profile of hCry2 was similar to that of hPer3. PMID: 14750904
- The carboxyl-terminal domain of human cryptochrome 2 is intrinsically unstructured in solution, consistent with its presumed role as a signal transduction domain. PMID: 15751956
- A clear circadian rhythm was shown for hPer1, hPer2, and hCry2 expression in CD34(+) cells and for hPer1 in the whole bone marrow, with maxima from early morning to midday. PMID: 17440215
- The expression level of CRY2 was decreased in hepatrocellular carcinoma. PMID: 18444243
- Both genetic association and functional analyses suggest that the circadian gene CRY2 may play an important role in NHL development. PMID: 19318546
显示更多
收起更多
-
亚细胞定位:Cytoplasm. Nucleus. Note=Translocated to the nucleus through interaction with other Clock proteins such as PER2 or ARNTL.
-
蛋白家族:DNA photolyase class-1 family
-
组织特异性:Expressed in all tissues examined including fetal brain, fibroblasts, heart, brain, placenta, lung, liver, skeletal muscle, kidney, pancreas, spleen, thymus, prostate, testis, ovary, small intestine, colon and leukocytes. Highest levels in heart and skele
-
数据库链接:
HGNC: 2385
OMIM: 603732
KEGG: hsa:1408
STRING: 9606.ENSP00000406751
UniGene: Hs.532491
Most popular with customers
-
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IF, FC
Species Reactivity: Human, Mouse, Rat
-
Phospho-YAP1 (S127) Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC
Species Reactivity: Human
-
-
-
-
-