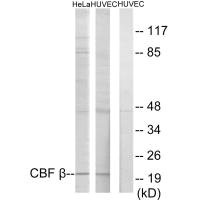
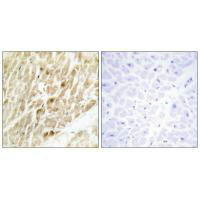
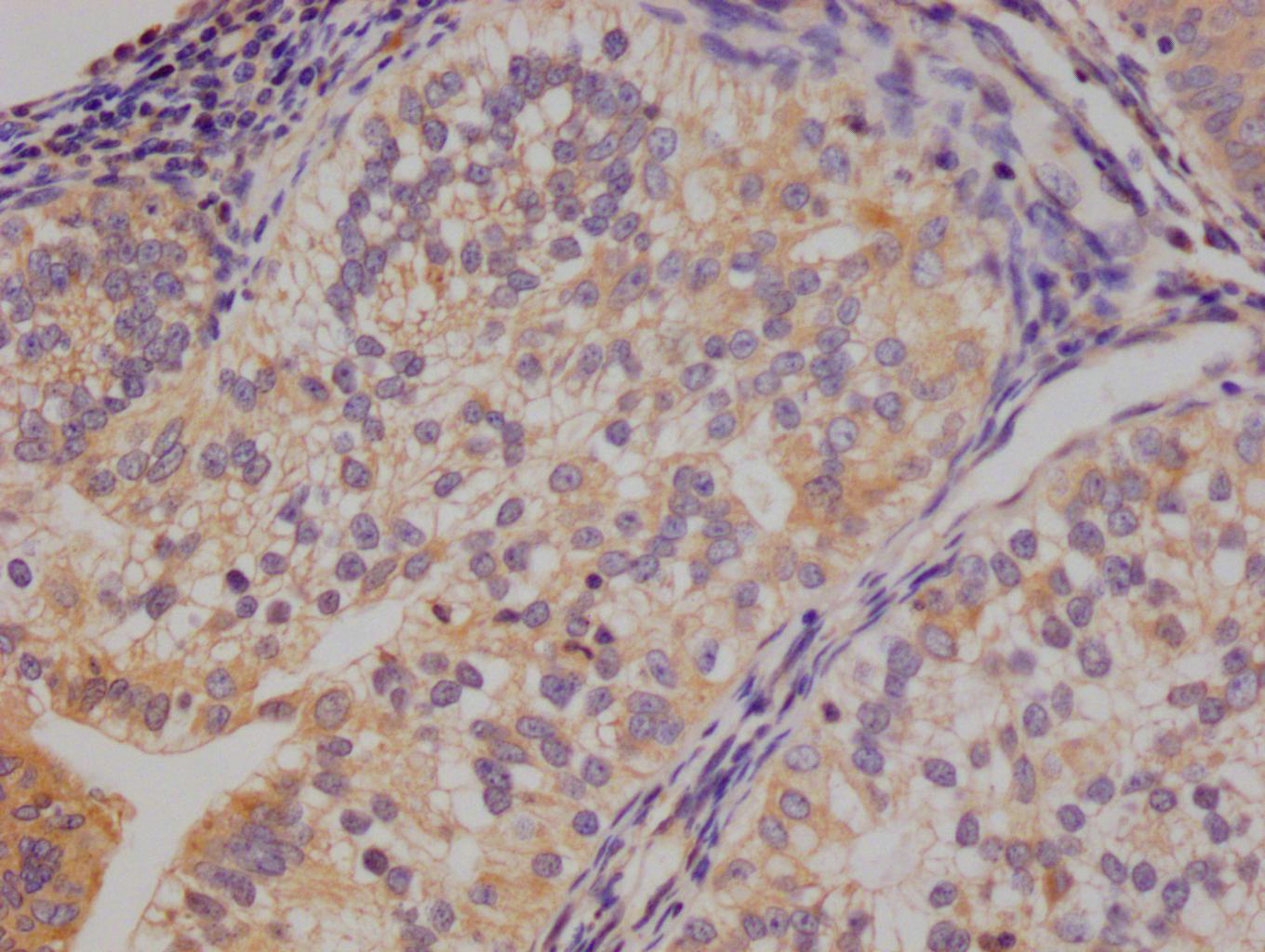
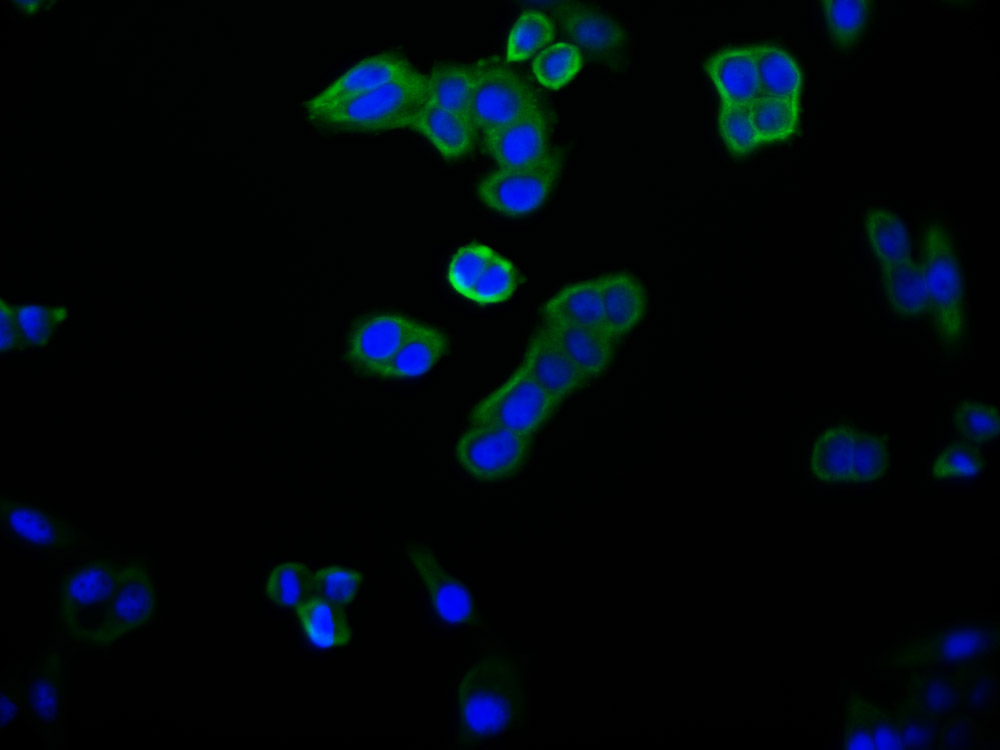
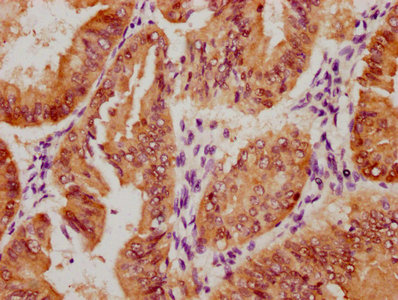
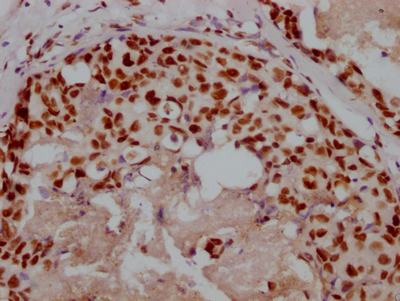
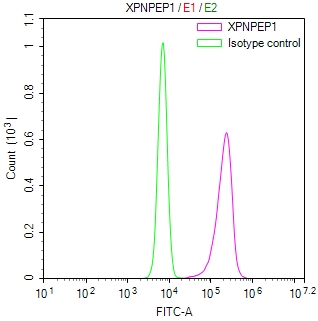
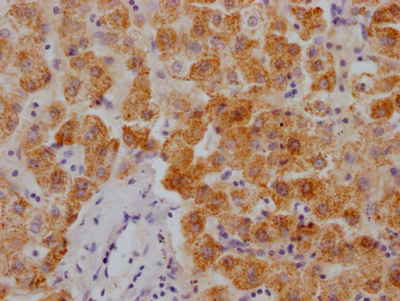
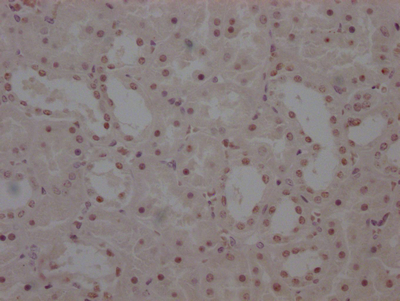
CBFB Antibody
-
货号:CSB-PA107678
-
规格:¥2024
-
图片:
-
其他:
产品详情
-
产品名称:Rabbit anti-Homo sapiens (Human) CBFB Polyclonal antibody
-
Uniprot No.:Q13951
-
基因名:
-
宿主:Rabbit
-
反应种属:Human,Mouse,Rat
-
免疫原:Synthesized peptide derived from internal of Human CBF β.
-
免疫原种属:Homo sapiens (Human)
-
克隆类型:Polyclonal
-
纯化方式:The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using epitope-specific immunogen.
-
浓度:It differs from different batches. Please contact us to confirm it.
-
产品提供形式:Liquid
-
应用范围:ELISA,WB,IHC
-
推荐稀释比:
Application Recommended Dilution WB 1:500-1:3000 IHC 1:50-1:100 -
Protocols:
-
储存条件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
货期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
相关产品
靶点详情
-
功能:Forms the heterodimeric complex core-binding factor (CBF) with RUNX family proteins (RUNX1, RUNX2, and RUNX3). RUNX members modulate the transcription of their target genes through recognizing the core consensus binding sequence 5'-TGTGGT-3', or very rarely, 5'-TGCGGT-3', within their regulatory regions via their runt domain, while CBFB is a non-DNA-binding regulatory subunit that allosterically enhances the sequence-specific DNA-binding capacity of RUNX. The heterodimers bind to the core site of a number of enhancers and promoters, including murine leukemia virus, polyomavirus enhancer, T-cell receptor enhancers, LCK, IL3 and GM-CSF promoters. CBF complexes repress ZBTB7B transcription factor during cytotoxic (CD8+) T cell development. They bind to RUNX-binding sequence within the ZBTB7B locus acting as transcriptional silencer and allowing for cytotoxic T cell differentiation.
-
基因功能参考文献:
- This study investigated the role of circ-CBFB in chronic lymphocytic leukemia. The ID of circ-CBFB in circBase is hsa_circ_0000707, which locates at chromosome 16q22.1 and derived from the back-splicing of CBFB transcript. PMID: 29902450
- results suggest that CBFbeta-SMMHC has complex actions on human ribosome biogenesis at both the genomic and posttranscriptional level PMID: 28196984
- the presented study demonstrates that CBFB-MYH11-based MRD status during the first 3 months after allo-HCT, but not KIT mutations, can be used to identify patients with a high risk of relapse. PMID: 27650511
- discussion of the role of CBFB in diseases caused by their mutations or deletions (review) PMID: 28299663
- Both c-kit receptor (KIT) D816V and KIT N822K mutations underwent autophosphorylation in the absence of growth factor in leukemia TF-1 cell line. PMID: 28506695
- The co-existence of BCR-ABL1 and CBFB rearrangement is associated with poor outcome and a clinical course similar to that of CML-BP, and unlike de novo AML with CBFB rearrangement, suggesting that high-intensity chemotherapy with TKI should be considered in these patients. PMID: 28253536
- Moreover, using a CBF-beta loss-of-function mutant, the s demonstrated that the interaction between CBF-beta and Vif was not sufficient for Vif assistance; a region including F68 in CBF-beta was also required for the stability and function of Vif. PMID: 28516844
- Vif stabilization by CBFbeta is mainly caused by impairing MDM2-mediated degradation. PMID: 27758855
- Mutational analysis of CBFbeta revealed that F68 and I55 residues are important and participate in a tripartite hydrophobic interaction with W5 of Vif to maintain a stable and functional Vif-CBFbeta complex. PMID: 28302150
- Our findings demonstrate that HSPCs exposed to non-cytotoxic levels of environmental chemicals and chemotherapeutic agents are prone to topoisomerase II-mediated DNA damage at the leukemia-associated genes MLL and CBFB. PMID: 26163765
- These results provide important information on the assembly of the Vif-CUL5-E3 ubiquitin ligase and identify a new viV binding interface with CBF-beta at the C-terminus of HIV-1 Vif. PMID: 25424878
- CBF-beta promoted steady-state levels of HIV-1 Vif by inhibiting the degradation of HIV-1 Vif through the proteasome pathway. PMID: 25582776
- CBFB contributes to the transcriptional regulation of ribosomal gene expression and provide further understanding of the epigenetic role of CBFB-SMMHC in proliferation and maintenance of the leukemic phenotype. PMID: 25079347
- we report a novel hypomethylation pattern, specific to CBFB-MYH11 fusion resulting from inv(16) rearrangement in acute myeloid leukemia the expression of which correlated with PBX3 differential methylation PMID: 25266220
- suggest that a different mechanism exists for the Vif-APOBEC interaction and that non-primates are not suitable animal models for exploring pharmacological interventions that disrupt Vif-CBF-beta interaction PMID: 25122780
- Suggest that CBFbeta retention in the midbody during cytokinesis reflects a novel function that contributes to epigenetic control. PMID: 24648201
- Transcriptional analysis revealed that upon fusion protein knockdown, a small subset of the CBFbeta-MYH11 target genes show increased expression, confirming a role in transcriptional repression PMID: 24002588
- s propose that CBFbeta acts as a chaperone to stabilize HIV-1 Vif during and after synthesis and to facilitate interaction of Vif with cellular cofactors required for the efficient degradation of APOBEC3G. PMID: 24522927
- In the absence of CBFbeta, Vif does not bind Cul5, thus preventing the assembly of the E3 ligase complex. PMID: 24390320
- CBF-beta is critical for the formation of the Vif-ElonginB/ElonginC-Cul5 core E3 ubiquitin ligase complex. PMID: 24390335
- Vif conserved residues E88/W89 are crucial for CBFbeta binding. PMID: 24418540
- data reveal the structural basis for Vif hijacking of the CBF-beta and CUL5 E3 ligase complex, laying a foundation for rational design of novel anti-HIV drugs PMID: 24402281
- This report of recurring FLT3 N676 mutations in core-binding factor (CBF) leukemias suggests a defined subgroup of CBF leukemias. PMID: 23878140
- We conclude that non-type A CBFB-MYH11 fusion types associate with distinct clinical and genetic features, including lack of KIT mutations, and a unique gene-expression profile in acute myeloid leukemia PMID: 23160462
- Our data indicate that the CBFbeta-SMMHC's C-terminus is essential to induce embryonic hematopoietic defects and leukemogenesis. PMID: 23152542
- A comparison of heat capacity changes supports a model in which CBFbeta prestabilizes Vif((1-192)) relative to Vif((95-192)) PMID: 23098073
- Vif proteins of human and simian immunodeficiency viruses require cellular CBFbeta to degrade APOBEC3G. PMID: 22205746
- Vif and CBF-beta physically interact, and that the amino-terminal region of Vif is required for this interaction PMID: 22190036
- CBF-beta is required for Vif-mediated degradation of APOBEC3G and therefore for preserving HIV-1 infectivity PMID: 22190037
- For routine clinical practice, it may be meaningful to screen for C-KIT mutations in AML1/ETO-positive patients, as well as for FLT3(D835) mutations in CBF-AML. PMID: 19603346
- The expression of Cbfbeta which were the key factors in osteogenic differentiation was also up-regulated. PMID: 20433876
- conclude that CBFbeta is required for a subset of Runx2-target genes that are sufficient to maintain the invasive phenotype of the cells PMID: 20591170
- Data collectively suggest that CBFbeta is required for malignant phenotype in prostate and ovarian cancer cells. PMID: 20607802
- has a role in hematopoiesis; preturbations result from expression of the leukemogenic fusion gene Cbfb-MYH11 PMID: 12239155
- Expression of CBFB is down regulated in a significant portion of gastric cancer cases; may be involved in gastric carcinogenesis PMID: 15386419
- Plag1 and Plagl2 are novel leukemia oncogenes that act by expanding hematopoietic progenitors expressing CbF beta-SMMHC. PMID: 15585652
- Detection of acute myeloid leukemic cells that are characterized by a CBFB-MYH11 gene fusion. PMID: 16502584
- These observations suggest that when abdominal GS is diagnosed, an analysis of the CBFB/MYH11 fusion gene is necessary to make an appropriate decision regarding treatment options, even if no chromosomal abnormalities are found. PMID: 16504290
- Agents interacting with the outer surface of the CBFbeta-SMMHC ACD that prevent multimerization may be effective as novel therapeutics in AML PMID: 16767164
- Rare fusion transcripts were correlated with an atypical cytomorphology not primarily suggestive for the FAB subtype acute myelocytic leukemia. PMID: 17287858
- Examine consequences of expression of abnormal chimeric protein CBFbeta-MYH11 in acute myelomonocytic leukemia. PMID: 17571080
- high CBFB protein level was an independent predictor of survival in colorectal cancer PMID: 19156145
显示更多
收起更多
-
相关疾病:A chromosomal aberration involving CBFB is associated with acute myeloid leukemia of M4EO subtype. Pericentric inversion inv(16)(p13;q22). The inversion produces a fusion protein that consists of the 165 N-terminal residues of CBF-beta (PEPB2) with the tail region of MYH11.
-
亚细胞定位:Nucleus.
-
蛋白家族:CBF-beta family
-
数据库链接:
HGNC: 1539
OMIM: 121360
KEGG: hsa:865
STRING: 9606.ENSP00000415151
UniGene: Hs.460988
Most popular with customers
-
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IF, FC
Species Reactivity: Human, Mouse, Rat
-
Phospho-YAP1 (S127) Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC
Species Reactivity: Human
-
-
-
-
-