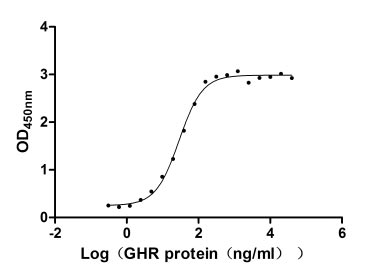
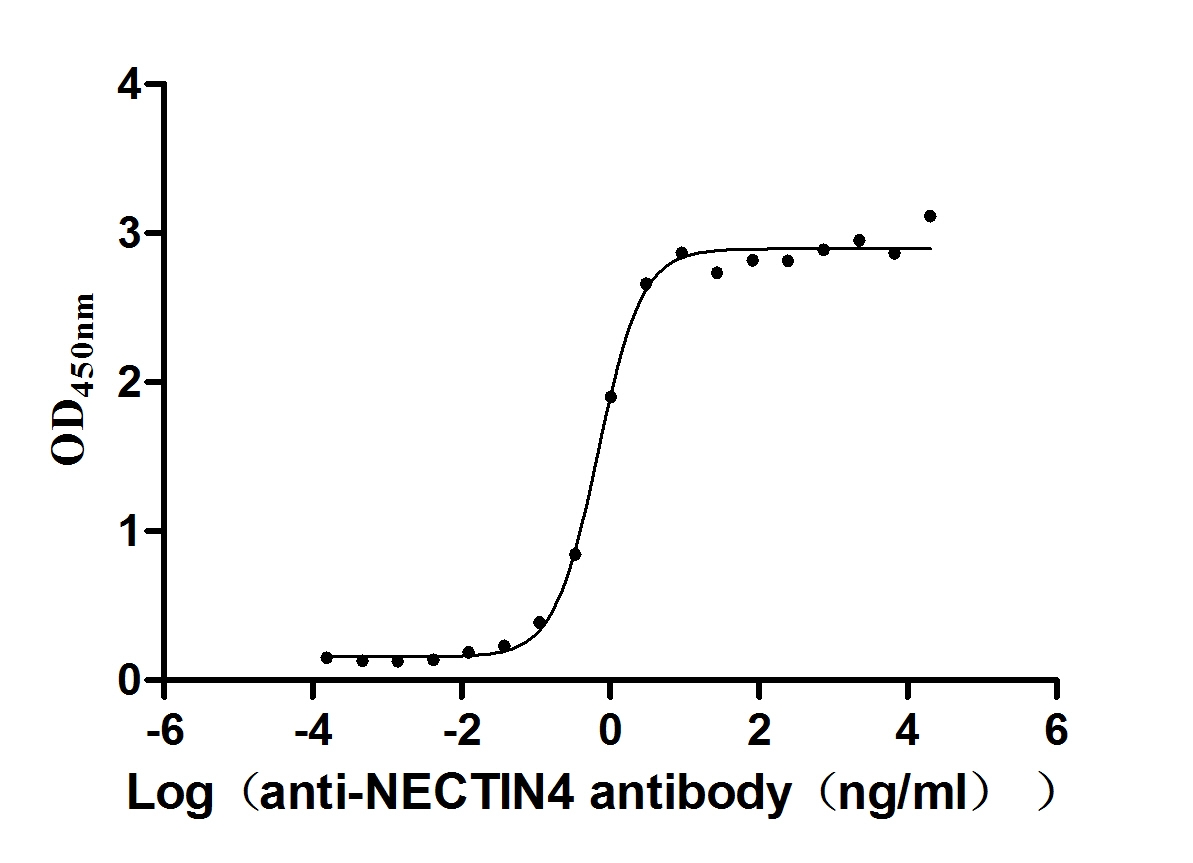
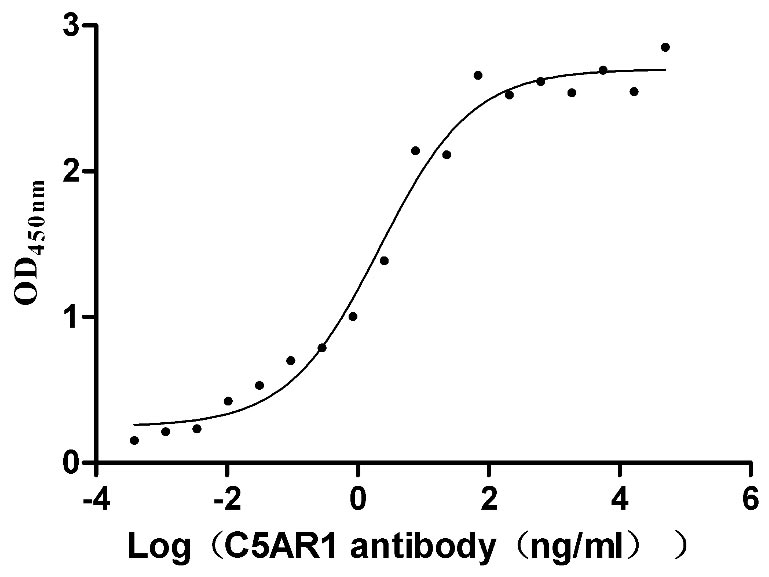
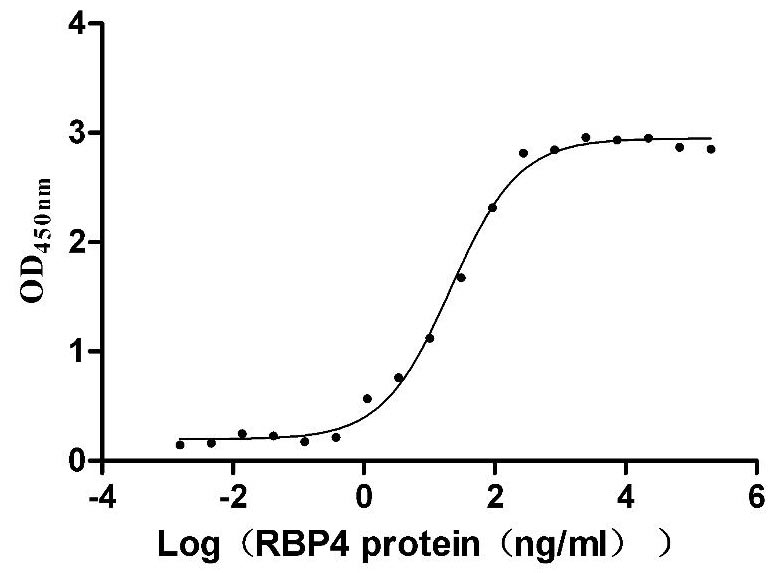
Recombinant Human Tumor necrosis factor receptor superfamily member 14 (TNFRSF14), partial
-
货号:CSB-YP842173HU
-
规格:
-
来源:Yeast
-
其他:
-
货号:CSB-EP842173HU
-
规格:
-
来源:E.coli
-
其他:
-
货号:CSB-EP842173HU-B
-
规格:
-
来源:E.coli
-
共轭:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
货号:CSB-BP842173HU
-
规格:
-
来源:Baculovirus
-
其他:
产品详情
-
纯度:>85% (SDS-PAGE)
-
基因名:
-
Uniprot No.:
-
别名:HVEML; ATAR; CD270; CD40 like protein precursor; Herpes virus entry mediator A; Herpesvirus entry mediator A; Herpesvirus entry mediator; Herpesvirus entry mediator ligand; HveA; HVEM; HVEM L; LIGHT; LIGHTR; TNFRSF14; TNFSF 14; TNR14_HUMAN; TR2; Tumor necrosis factor receptor like gene2; Tumor necrosis factor receptor superfamily member 14; Tumor necrosis factor receptor superfamily member 14 precursor; Tumor necrosis factor receptor-like 2
-
种属:Homo sapiens (Human)
-
蛋白长度:Partial
-
蛋白标签:Tag type will be determined during the manufacturing process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
产品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
相关产品
靶点详情
-
功能:Receptor for four distinct ligands: The TNF superfamily members TNFSF14/LIGHT and homotrimeric LTA/lymphotoxin-alpha and the immunoglobulin superfamily members BTLA and CD160, altogether defining a complex stimulatory and inhibitory signaling network. Signals via the TRAF2-TRAF3 E3 ligase pathway to promote immune cell survival and differentiation. Participates in bidirectional cell-cell contact signaling between antigen presenting cells and lymphocytes. In response to ligation of TNFSF14/LIGHT, delivers costimulatory signals to T cells, promoti...显示更多
-
基因功能参考文献:
- Data suggest that both HVEM and UL144 bind a common epitope of BTLA, whether engaged in trans or in cis; these studies were conducted in cell lines representing B-lymphocytes, T-lymphocytes, and natural killer cells. (HVEM = human herpes virus entry mediator; UL144 = membrane glycoprotein UL144 of Human herpesvirus 5; BTLA = human B- and T-lymphocyte attenuator) PMID: 29061848
- our data suggested that the BTLA/HVEM pathway contributes to peripheral T cell suppression in hepatocellular carcinoma patients PMID: 30116751
- TNFRSF14 may serve a tumor suppressive role in bladder cancer by inducing apoptosis and suppressing proliferation, and act as a novel prognostic biomarker for bladder cancer. PMID: 30066919
- Primary cutaneous follicle center lymphomas with concomitant 1p36 deletion and TNFRSF14 mutations frequently express high levels of EZH2 protein. PMID: 29858685
- High HVEM Expression is Associated with Cancer Progression in Breast Cancer. PMID: 28612127
- Report a variant of t(14;18) negative nodal diffuse follicular lymphoma with CD23 expression, 1p36/TNFRSF14 abnormalities, and STAT6 mutations. PMID: 26965583
- Roles of HVEM are likely to be immunosuppressive rather than activating tumor immunity and it in peripheral blood is a diagnostic marker and therapeutic target for hepatocellular carcinoma. PMID: 27987232
- Low HVEM expression is associated with pancreatic and ampullary cancer. PMID: 28470686
- HIV-1 produced from CD4+ T cells bears HSV-2 receptor HVEM and can bind to and enter HSV-2-infected epithelial cells depending on HVEM-gD interaction and the presence of gB/gH/gL. PMID: 28809154
- Transgenic mice expressing HVEMIg showed a complete resistance to the lethal infection even with 300 MLD50 (survival rate of 100 %). PMID: 28671524
- HVEM is highly expressed in ovarian serous adenocarcinoma tissues and correlated with the patient clinicopathological features. PMID: 28365939
- TNFRSF14 and MAP2K1 mutations are the most frequent genetic alterations found in pediatric-type follicular lymphoma (PTFL) and occur independently in most cases, suggesting that both mutations might play an important role in PTFL lymphomagenesis. PMID: 28533310
- genetic landscape of Pediatric-type follicular lymphoma suggests that TNFRSF14 mutations accompanied by copy-number neutral loss of heterozygosity of the 1p36 locus in over 70% of mutated cases, as additional selection mechanism, might play a key role in the pathogenesis of this disease. PMID: 27257180
- The increased immune-stimulatory capacity of lymphoma B cells with TNFRSF14 aberrations had clinical relevance, associating with higher incidence of acute GVHD in patients undergoing allogeneic hematopoietic stem cell transplantation. PMID: 27103745
- These results suggest that TNFRSF14 mutations point towards a diagnosis of follicular lymphomas , and can be used in the sometimes difficult distinction between marginal zone lymphomas and follicular lymphomas PMID: 27297871
- the overexpression of HVEM in ovarian cancer cells may suppress the proliferation and immune function of T cells, thus leading to the development of ovarian cancer. The current study partially explains the immune escape mechanism of ovarian cancer cells. PMID: 27458100
- In eight cases (42%) we observed recurrent copy number loss of chr1:2,352,236-4,574,271, a region containing the candidate tumor suppressor TNFRSF14. PMID: 26650888
- Study report the crystal structure of unbound HVEM, which further contributes to the understanding of the molecular mechanisms controlling recognition between HVEM and its ligands. PMID: 26202493
- HVEM may play a critical role in tumor progression and immune evasion PMID: 25750286
- Data indicate that tumour-expressing herpes virus entry mediator (HVEMplays a critical role in hepatocellular carcinoma (HCC), suggesting targeting HVEM may be a promising therapeutic strategy for HCC. PMID: 25468715
- Relative expression of HVEM and LTbetaR modulates canonical NF-kappaB and pro-apoptotic signals stimulated by LIGHT. PMID: 24980868
- Sequencing of TNFRSF14 located in the minimal region of loss in 1p36.32 showed nine mutations in pediatric follicular lymphoma. PMID: 23445872
- HVEM plays a critical role in both tumor progression and the evasion of host antitumor immune responses, possibly through direct and indirect mechanisms. PMID: 24249528
- HVEM gene polymorphisms are associated with sporadic breast cancer in Chinese women. PMID: 23976978
- The conformation of the N-terminus of herpes simplex virus gD is induced by direct binding to HVEM and nectin-1. PMID: 24314649
- HVEM functions as a regulator of immune function that activates NK cells via CD160 and limits lymphocyte-induced inflammation via association with B and T lymphocyte attenuator PMID: 23761635
- BTLA and HVEM may have roles in graft rejection after kidney transplantation PMID: 23375291
- Studies indicate co-stimulatory and co-inhibitory receptors B7-1, B7-2, CD28 and TNFRSF14 have a pivotal role in T cell biology, as they determine the functional outcome of T cell receptor (TCR) signalling. PMID: 23470321
- These findings support role for BTLA and/or HVEM as potential, novel diagnostic markers of innate immune response/status and as therapeutic targets of sepsis. PMID: 22459947
- study described the expression and spatial distribution of HVEM and BTLA in rheumatoid arthritis synovial tissues, and results indicated that HVEM/BTLA may be involved in regulating the progress of joint inflammation PMID: 22179929
- HVEM-B and T lymphocyte attenuator (BTLA) interactions impair minor histocompatibility antigen (MiHA)-specific T cell functionality, providing a rationale for interfering with BTLA signaling in post-stem cell transplantation. PMID: 22634623
- Results indicate that mHVEM on leukocytes and sHVEM in sera may contribute to the development and/or progression of gastric cancer. PMID: 22113134
- These results suggest that the C-terminal portion of the soluble HVEM ectodomain inhibits herpes simplex virus type 1 gD activation and that this effect is neutralized in the full-length form of HVEM in normal infection. PMID: 22239829
- TNFRSF14 appears to be a serious candidate gene that might contribute to follicular lymphoma development. PMID: 21941365
- HVEM-BTLA cis complex provides intrinsic regulation in T cells serving as an interference mechanism silencing signals coming from the microenvironment. PMID: 21920726
- The results of a mutagenesis study of HVEM suggest that the CD160 binding region on HVEM was slightly different from, but overlapped with, the BTLA binding site. PMID: 21959263
- data show that HVEM stimulatory signals promote experimental colitis driven by innate or adaptive immune cells PMID: 21533159
- Polymorphisms were associated with MS predisposition, with stronger effect in patients with HHV6 active replication-TNFRSF6B-rs4809330(*)A: P=0.028, OR=1.13; TNFRSF14-rs6684865(*)A: overall P=0.0008, OR=1.2. PMID: 20962851
- Findings identify TNFRSF14 as a candidate gene associated with a subset of FL, based on frequent occurrence of acquired mutations and their correlation with inferior clinical outcomes. PMID: 20884631
- We have identified and replicated a novel gene-gene interaction between 2 polymorphisms of TNFRSF members in Spanish patients with RA, based on the hypothesis of shared pathogenic pathways in complex diseases. PMID: 20187130
- Results provide evidence of an existing relationship between HVEM and obesity, which suggest that this TNF superfamily receptor could be involved in the pathogenesis of obesity and inflammation-related activity. PMID: 19680232
- Data suggest involvement of TNF superfamily receptor members and ligands in human atherosclerosis. TNFRSF14 (HVEM, TR2, LIGHTR)analysis, found this receptor in regions rich in CD68-positive macrophage-derived foam cells and HLA-DR-positive cells. PMID: 11742858
- Crystallization and preliminary diffraction studies of the ectodomain of the envelope glycoprotein D from herpes simplex virus 1 alone and in complex with the ectodomain of the human receptor HveA PMID: 11976496
- association of HVEM and nectin-1 with lipid rafts during herpes simplex virus entry PMID: 12915568
- sHVEM levels were elevated in sera of patients with allergic asthma, atopic dermatitis and rheumatoid arthritis PMID: 14749527
- both nectin 1 and HVEM receptors play a role during HSV infection in vivo and both are highly efficient even at low levels of expression PMID: 15110526
- Binding of HVEM to BTLA attenuates T cell activation, identifying HVEM/BTLA as a coinhibitory receptor pair. PMID: 15647361
- in cells a complex forms through physical associations of HVEM, HSV-1 gD, and at least gH PMID: 15767456
- distinct herpesviruses target the HVEM-BTLA cosignaling pathway, suggesting the importance of this pathway in regulating T cell activation during host defenses. PMID: 16131544
- 2.8-A crystal structure of the BTLA-HVEM complex shows that BTLA binds the N-terminal cysteine-rich domain of HVEM and employs a unique binding surface PMID: 16169851
收起更多
-
亚细胞定位:Cell membrane; Single-pass type I membrane protein.
-
组织特异性:Widely expressed, with the highest expression in lung, spleen and thymus. Expressed in a subpopulation of B cells and monocytes. Expressed in naive T cells.
-
数据库链接:
HGNC: 11912
OMIM: 602746
KEGG: hsa:8764
STRING: 9606.ENSP00000347948
UniGene: Hs.512898