Recombinant Human Purine nucleoside phosphorylase (PNP)
-
货号:CSB-YP355527HU
-
规格:
-
来源:Yeast
-
其他:
-
货号:CSB-EP355527HU-B
-
规格:
-
来源:E.coli
-
共轭:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
货号:CSB-BP355527HU
-
规格:
-
来源:Baculovirus
-
其他:
-
货号:CSB-MP355527HU
-
规格:
-
来源:Mammalian cell
-
其他:
产品详情
-
纯度:>85% (SDS-PAGE)
-
基因名:PNP
-
Uniprot No.:
-
别名:FLJ94043; FLJ97288; FLJ97312; Inosine phosphorylase; Inosine-guanosine phosphorylase; MGC117396; MGC125915; MGC125916; NP; Np1; Nucleoside phosphorylase; PNP; Pnp1; PNPH_HUMAN; PRO1837; PUNP; Purine nucleoside orthophosphate ribosyltransferase; Purine nucleoside phosphorylase 5a; Purine nucleoside phosphorylase
-
种属:Homo sapiens (Human)
-
蛋白长度:full length protein
-
表达区域:1-289
-
氨基酸序列MENGYTYEDY KNTAEWLLSH TKHRPQVAII CGSGLGGLTD KLTQAQIFDY GEIPNFPRST VPGHAGRLVF GFLNGRACVM MQGRFHMYEG YPLWKVTFPV RVFHLLGVDT LVVTNAAGGL NPKFEVGDIM LIRDHINLPG FSGQNPLRGP NDERFGDRFP AMSDAYDRTM RQRALSTWKQ MGEQRELQEG TYVMVAGPSF ETVAECRVLQ KLGADAVGMS TVPEVIVARH CGLRVFGFSL ITNKVIMDYE SLEKANHEEV LAAGKQAAQK LEQFVSILMA SIPLPDKAS
-
蛋白标签:Tag type will be determined during the manufacturing process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
产品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
相关产品
靶点详情
-
功能:Catalyzes the phosphorolytic breakdown of the N-glycosidic bond in the beta-(deoxy)ribonucleoside molecules, with the formation of the corresponding free purine bases and pentose-1-phosphate. Preferentially acts on 6-oxopurine nucleosides including inosine and guanosine.
-
基因功能参考文献:
- The study suggests that mass-constrained femtosecond motions at the catalytic site of PNP can improve transition state barrier crossing by more frequent sampling of essential catalytic site contacts. PMID: 29915028
- The PNP rs1049564 T allele is a loss-of-function variant that induces S-phase block and IFN pathway activation in lymphocytes. The S-phase block could be rescued in our in vitro experiments, suggesting the potential for personalized treatment. PMID: 28859258
- Data show that the mutations in purine nucleoside phosphorylase (PNP) alters the enthalpy-entropy balance with little effect on the catalytic rates. PMID: 27976868
- Data (including data from empirical valence bond/molecular dynamic simulations) suggest that PNP substrate specificity for inosine and guanosine is a direct result of electrostatic preorganization energy along the reaction coordinate. PMID: 26985580
- the binding mechanism of a transition state analogue (DADMe-immucillin-H) to the purine nucleoside phosphorylase (PNP) enzyme, is reported. PMID: 25625196
- Data show that [15N, 2H]His8-purine nucleoside phosphorylase (PNP) had reduced catalytic site chemistry larger than proportional to the enzymatic mass difference. PMID: 26305965
- Study of genetic heterogeneity in systemic lupus erythematosus, the top associations in European ancestry were protein kinase, cyclic GMP-dependent, type I (PRKG1) rs7897633 (P(Meta) = 2.75 x 10(-8)) and purine nucleoside phosphorylase (PNP) rs1049564 (P(Meta) = 1.24 x 10(-7)). PMID: 25338677
- Human small intestine is a key site for ribavirin phosphorolysis and that PNP is primarily involved in the metabolism. PMID: 24107682
- insufficient data to evaluated impact of genetic polymorphisms on disease susceptibility PMID: 24792412
- Complete lack of PNP triggers accumulation of deoxyguanosine, thereby disrupting B-cell development, the consequence of which is more profound with time, as was found in the older sister. PMID: 22578971
- Biochemical and genetic data on a cohort of seven patients from six families identified as PNPase deficient, is reported. PMID: 22132981
- This study for the first time describes elevated levels of alpha synuclein in pancreatic adenocarcinoma as well as highlights the potential of evaluating NP protein expression. PMID: 21448452
- investigation of catalytic mechanisms involved in catalysis by PNP: transition states in arsenolysis and phosphorolysis PMID: 21348499
- Results show that some regions, responsible for entrance and exit of substrate, present a conformational variability, which is dissected by dynamics simulation analysis. PMID: 19932753
- PNP operating at maximum catalytic potential permits more rapid peptide amide deuterium exchange and greater conformational flexibility of water-peptide bond exchange rate than in either of the complexes with transition state analogues. PMID: 20108972
- The optimum pH for PNP from human erythrocytes with xanthosine and xanthine is in the range 5-6, whereas those with guanosine, guanine, inosine & hypoxanthine are in the range 7-8. Possible PNP binding modes of Xan and Xao by mammalian PNPs are proposed. PMID: 12180982
- Crystal structure of human purine nucleoside phosphorylase. PMID: 12914785
- These data provide a framework in which to conduct genetic association studies of these two genes in relevant populations, thereby allowing hNP and hGSTO1-1 to be evaluated as potential susceptibility genes in human arsenicism. PMID: 12928150
- investigation of the quaternary structure of recombinant human purine nucleoside phosphorylase PMID: 13679062
- crystal structures in complex with inosine and 2',3'-dideoxyinosine, refined to 2.8A resolution using synchrotron radiation. The structures provide explanation for ligand binding, refine the purine-binding site and can be used for future inhibitor design. PMID: 14706628
- several recurring mutations were found in PNP in patients with purine nucleoside phosphorylase deficiency by DNA sequence analysis PMID: 15571269
- crystal structure of human PNP in complex with hypoxanthine, refined to 2.6A resoluti PMID: 15582582
- findings suggest that the G51S PNP polymorphism is associated with a faster rate of cognitive decline in Alzheimer's disease patients, highlighting the important role of purine metabolism in the progression of this neurodegenerative disorder PMID: 17221831
- Role of ionization of the phosphate cosubstrate on phosphorolysis by purine nucleoside phosphorylase PMID: 17639373
- Altered thermodynamics from remote mutations altering human toward bovine purine nucleoside phosphorylase. PMID: 18281956
- New interactions caused by the mutations increase the catalytic efficiency of the enzyme for formation of a late transition state with increased participation of the phosphate nucleophile. PMID: 18281957
- Structural studies on NP are reported with a view towards a new specific scoring function. PMID: 18790691
- Protein dynamics on the femtosecond to picosecond timescale are linked to enzymatic function. PMID: 18946041
- Comparative analysis of the model of BfPNP and the structure of HsPNP allowed identification of structural features responsible for differences in the computationally determined ligand affinities PMID: 19172318
- Results describe a tryptophan-free mutant of purine nucleoside phosphorylase and its dynamic activity. PMID: 19191546
- Altered enthalpy-entropy compensation in picomolar transition state analogues of human purine nucleoside phosphorylase PMID: 19425594
显示更多
收起更多
-
相关疾病:Purine nucleoside phosphorylase deficiency (PNPD)
-
亚细胞定位:Cytoplasm.
-
蛋白家族:PNP/MTAP phosphorylase family
-
组织特异性:Expressed in red blood cells; overexpressed in red blood cells (cytoplasm) of patients with hereditary non-spherocytic hemolytic anemia of unknown etiology.
-
数据库链接:
HGNC: 7892
OMIM: 164050
KEGG: hsa:4860
STRING: 9606.ENSP00000354532
UniGene: Hs.75514
Most popular with customers
-
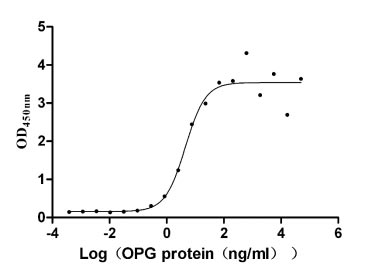
Recombinant Human Tumor necrosis factor receptor superfamily member 11B (TNFRSF11B) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Intestinal-type alkaline phosphatase (ALPI) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
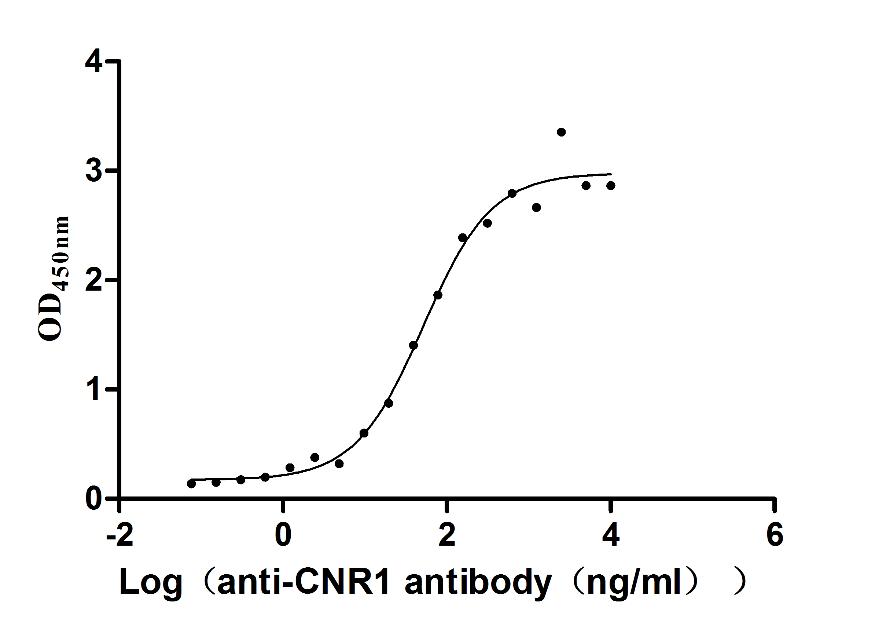
Recombinant Human Cannabinoid receptor 1 (CNR1)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
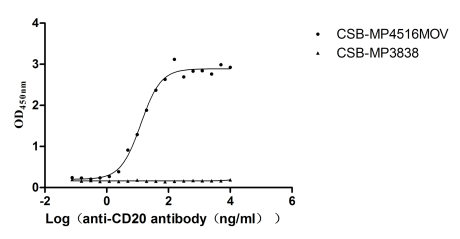
Recombinant Macaca fascicularis Membrane spanning 4-domains A1 (MS4A1)-VLPs (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
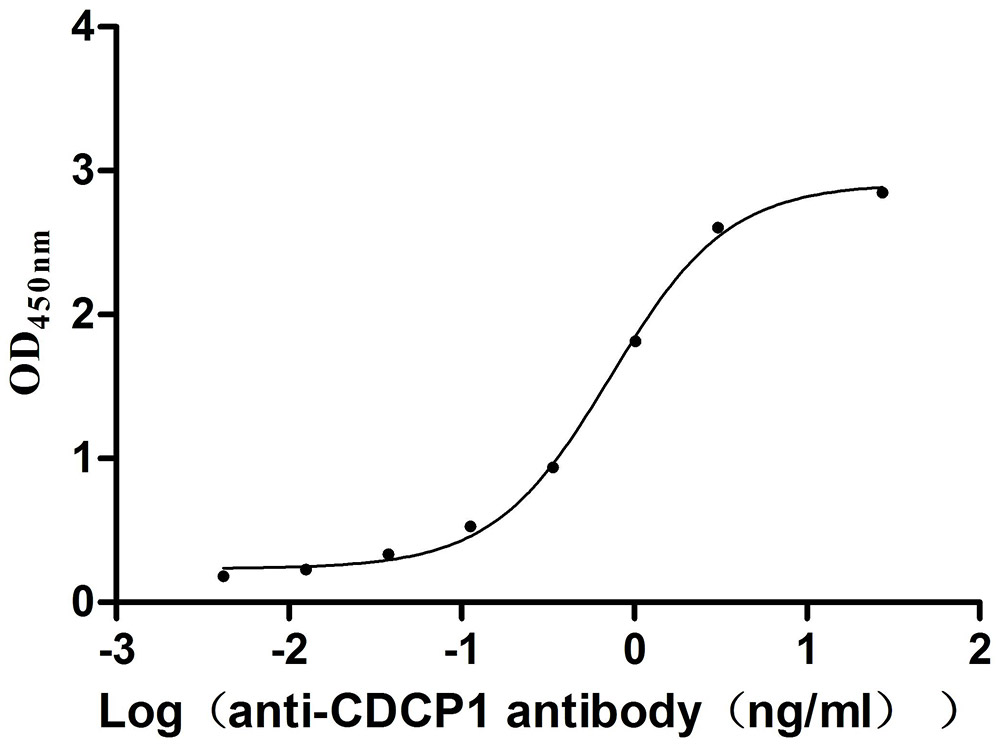
Recombinant Mouse CUB domain-containing protein 1 (Cdcp1), partial (Active)
Express system: Mammalian cell
Species: Mus musculus (Mouse)
-
Recombinant Human CD81 antigen (CD81), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)