Recombinant Human DNA mismatch repair protein Msh6 (MSH6), partial
-
货号:CSB-EP015032HU
-
规格:¥1344
-
图片:
-
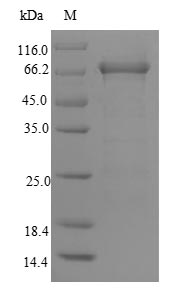
(Tris-Glycine gel) Discontinuous SDS-PAGE (reduced) with 5% enrichment gel and 15% separation gel.
-
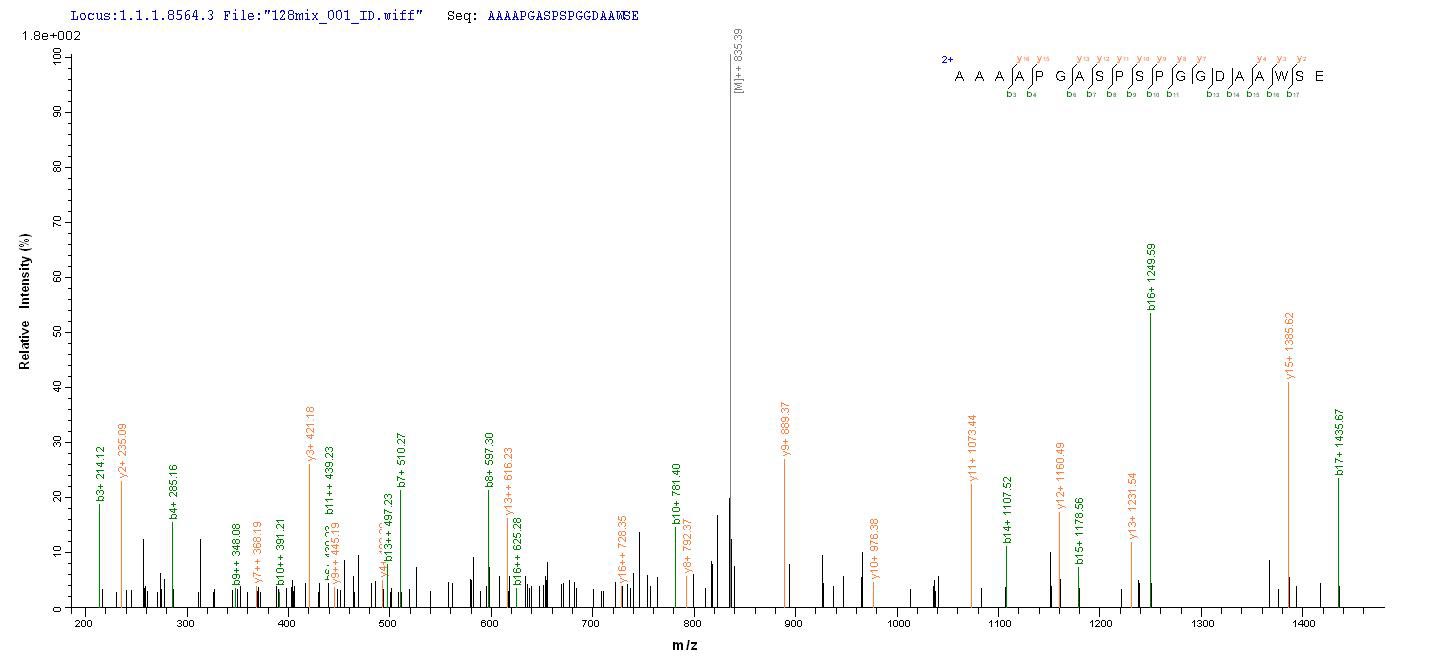
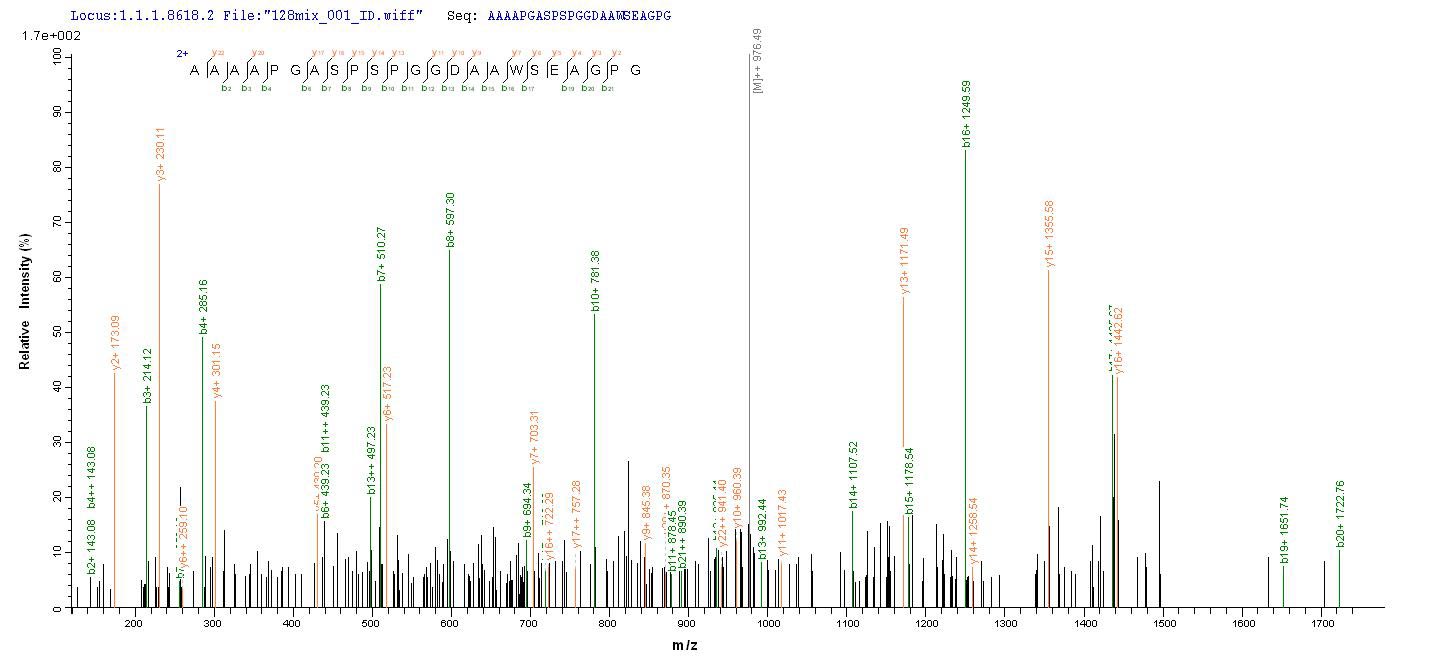
Based on the SEQUEST from database of E.coli host and target protein, the LC-MS/MS Analysis result of CSB-EP015032HU could indicate that this peptide derived from E.coli-expressed Homo sapiens (Human) MSH6.
-
Based on the SEQUEST from database of E.coli host and target protein, the LC-MS/MS Analysis result of CSB-EP015032HU could indicate that this peptide derived from E.coli-expressed Homo sapiens (Human) MSH6.
-
-
其他:
产品详情
-
纯度:Greater than 90% as determined by SDS-PAGE.
-
基因名:
-
Uniprot No.:
-
别名:DNA mismatch repair protein Msh6; G/T mismatch binding protein; G/T mismatch-binding protein; GTBP; GTMBP; hMSH6; HNPCC 5; HNPCC5; HSAP; MSH 6; MSH6; MSH6_HUMAN; mutS (E. coli) homolog 6; MutS alpha 160 kDa subunit; MutS homolog 6 (E. coli); mutS homolog 6; MutS-alpha 160 kDa subunit; p160; Sperm associated protein
-
种属:Homo sapiens (Human)
-
蛋白长度:Partial
-
来源:E.coli
-
分子量:60.1kDa
-
表达区域:1-400aa
-
氨基酸序列MSRQSTLYSFFPKSPALSDANKASARASREGGRAAAAPGASPSPGGDAAWSEAGPGPRPLARSASPPKAKNLNGGLRRSVAPAAPTSCDFSPGDLVWAKMEGYPWWPCLVYNHPFDGTFIREKGKSVRVHVQFFDDSPTRGWVSKRLLKPYTGSKSKEAQKGGHFYSAKPEILRAMQRADEALNKDKIKRLELAVCDEPSEPEEEEEMEVGTTYVTDKSEEDNEIESEEEVQPKTQGSRRSSRQIKKRRVISDSESDIGGSDVEFKPDTKEEGSSDEISSGVGDSESEGLNSPVKVARKRKRMVTGNGSLKRKSSRKETPSATKQATSISSETKNTLRAFSAPQNSESQAHVSGGGDDSSRPTVWYHETLEWLKEEKRRDEHRRRPDHPDFDASTLYVPE
Note: The complete sequence including tag sequence, target protein sequence and linker sequence could be provided upon request. -
蛋白标签:N-terminal 6xHis-SUMO-tagged
-
产品提供形式:Liquid or Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
缓冲液:Tris-based buffer,50% glycerol
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet & COA:Please contact us to get it.
引用文献
相关产品
靶点详情
-
功能:Component of the post-replicative DNA mismatch repair system (MMR). Heterodimerizes with MSH2 to form MutS alpha, which binds to DNA mismatches thereby initiating DNA repair. When bound, MutS alpha bends the DNA helix and shields approximately 20 base pairs, and recognizes single base mismatches and dinucleotide insertion-deletion loops (IDL) in the DNA. After mismatch binding, forms a ternary complex with the MutL alpha heterodimer, which is thought to be responsible for directing the downstream MMR events, including strand discrimination, excision, and resynthesis. ATP binding and hydrolysis play a pivotal role in mismatch repair functions. The ATPase activity associated with MutS alpha regulates binding similar to a molecular switch: mismatched DNA provokes ADP-->ATP exchange, resulting in a discernible conformational transition that converts MutS alpha into a sliding clamp capable of hydrolysis-independent diffusion along the DNA backbone. This transition is crucial for mismatch repair. MutS alpha may also play a role in DNA homologous recombination repair. Recruited on chromatin in G1 and early S phase via its PWWP domain that specifically binds trimethylated 'Lys-36' of histone H3 (H3K36me3): early recruitment to chromatin to be replicated allowing a quick identification of mismatch repair to initiate the DNA mismatch repair reaction.
-
基因功能参考文献:
- discover a translocation disrupting MLH1 and three mutations in MSH6 and PMS2 that increase endometrial, colorectal, brain and ovarian cancer risk PMID: 28466842
- Whole exome sequencing in triple negative breast cancer cases revealed MSH6 rs2020912 (MAF: 56.25% vs. 1.04%, OR 122.13, 95% CI 12.29-2985.48) are risk factors for triple negative breast cancer. PMID: 30136158
- CSE1L expression was correlated with MSH6 expression in tumor samples and was associated with poor prognosis in patients with osteosarcoma. Taken together, our results demonstrate that the CSE1L-MSH6 axis has an important role in osteosarcoma progression. PMID: 28387323
- our data suggests thatMSH6 Glu39Gly polymorphism is associated with the risk of developing sporadic colorectal cancer in polish population. Linkage to the female gender, onset above 60 years old and further increase of risk when combined with wild-type allele of PMS2 IVS1-1121C > PMID: 28451866
- Results from a study on gene expression variability markers in early-stage human embryos shows that is a putative expression variability marker for the 3-day, 8-cell embryo stage. PMID: 26288249
- hMSH6 Glu39Gly polymorphism is associated with the risk of developing colorectal cancer in the Polish population. PMID: 29442465
- Expression of MSH6 and MSH2 was positively associated with tumor volume doubling time. Gene expression was positively associated with ATR expression. Reduction of MSH6 and MSH2 expression at the messenger RNA and protein levels could be involved in direct Pituitary Adenoma proliferation by promoting cell-cycle progression or decreasing the rate of apoptosis through interference with the function of the ATR-Chk1 pathway. PMID: 29342268
- Our results suggest that increased expression of MSH6, or other MMR, may be a new mechanism contributing to the acquired resistance during TMZ therapy; and may serve as an indicator to the resistance in GBM. PMID: 29366782
- study to estimate frequency of point mutations and chromosomal rearrangements in 3 mismatch repair (MMR) genes MLH1, MSH2, and MSH6 among unselected patients with ovarian cancer; estimate that approximately 0.6% of unselected ovarian cancer patients have mutations in the MMR genes PMID: 28176205
- MSH6 frameshift variants incompletely segregate with the Lynch syndrome phenotype in two families. PMID: 28481244
- Patients with mutations 6 showed higher rate of achieving major molecular response than those<6 (P=0.0381). Mutations in epigenetic regulator, ASXL1, TET2, TET3, KDM1A and MSH6 were found in 25% of patients. TET2 or TET3, AKT1 and RUNX1 were mutated in one patient each. ASXL1 was mutated within exon 12 in three cases PMID: 28452984
- The MSH6 gene polymorphisms are likely to play an important role in the progression of AIDS in the northern Chinese population. PMID: 27090025
- MSH6 mutations contribute to colorectal cancer susceptibility in Algerian families with suspected Lynch syndrome. PMID: 27468915
- Neoadjuvant therapy in microsatellite-stable colorectal carcinoma induces concomitant loss of MSH6 and Ki-67 expression. PMID: 28232158
- High MSH6 expression is associated with the development of genetic instability and is linked to tumor aggressiveness and early PSA recurrence in prostate cancer. PMID: 27803051
- In path_MSH6 10-year survival was 84%, relative incidence of subsequent cancer compared with incidence of first cancer was slightly but insignificantly higher than cancer incidence in patients with Lynch syndrome without previous cancer. PMID: 27261338
- human Pol alpha interacts with MSH2-MSH6 complex PMID: 27805738
- In colorectal neoplasms, negative expression of the MMR proteins MLH1, MSH2 or MSH6 was seen in 15% (47 of 313) of the patients. Defect MLH1 was most common and detected in 12% of the cases. Defect MLH1 and MSH2 were identified in each patient's normal adjacent mucosa. PMID: 27836416
- A total of 201 unique disease-predisposing mismatch repair gene mutations were identified in 369 Lynch syndrome families. These mutations affected MLH1 in 40%, MSH2 in 36%, MSH6 in 18% and PMS2 in 6% of the families PMID: 27601186
- There is a positive correlation between the expressions of hMSH2 and hMSH6 between males (RHO=0.673 and p=0.001) and females with colorectal adenocarcinoma. PMID: 27459116
- Individuals with Lynch syndrome and double-mutants in MSH6 and MSH2 had normal MSH2 expression, whereas MSH6 immunoexpression was lost in all evaluable cases. PMID: 26446363
- we identified gene promoter methylation signatures (WT1, MSH6, GATA5 and PAX5) that are strongly correlated to, and can have a predictive value for the clinical outcome of oral squamous cell carcinoma patients PMID: 27491556
- In Lynch syndrome families, prostate cancer was associated with mutations in MSH6 with loss of the mismatch repair protein. PMID: 27013479
- Loss of MSH-2/MSH-6 expression was correlated with the right-colon location, poor and mucinous differentiation in colorectal adenocarcinoma. PMID: 26097592
- Heterozygous germline mutations in any of the mismatch repair (MMR) genes, MLH1, MSH2, MSH6, and PMS2, cause Lynch syndrome (LS), an autosomal dominant cancer predisposition syndrome. PMID: 26544533
- MSH2, MSH6, and EXO1 genes were overexpressed in gastroesophageal cancers. PMID: 26215063
- SNP rs1800934 and 12 rare variants in the mismatch repair gene MSH6 were also associated with cafe-au-lait macule count in neurofibromatosis type 1 patients. PMID: 25329635
- microsatellite instability-high and loss of MSH6 were found. Based on these results, genetic testing of MSH6 revealed a frame-shift mutation in codon 604 PMID: 26805314
- MutSalpha, proliferating cell nuclear antigen, and replication factor C activate MutLalpha endonuclease to remove the 1-nucleotide Okazaki fragment flaps PMID: 26224637
- We describe a kindred with multiple gastrointestinal malignancies where a novel MSH6 germline susceptibility variant was identified PMID: 25380764
- Heterogenous MSH6 loss is uncommon, usually caused by instability in MSH6 exon 5 polycytosine tract, and not associated with germline MSH6 mutation. PMID: 26099011
- immunohistochemical expression of MLH1, MSH2, and MSH6 in initial glioblastoma is not associated with patient survival PMID: 24995467
- The study evaluates the frequency and distribution of mutations in the MLH1, MSH2 and MSH6 genes within a cohort of families with Lynch syndrome in Cyprus. PMID: 25133505
- Somatic rearrangements in MSH2 and MSH6 are an important mechanism leading to hypermutation and microsatellite instability in advanced prostate cancer. PMID: 25255306
- Our database and literature searches retrieved 30 MLH1, 22 MSH2, 4 MSH6 and 9 PMS2 alternative transcripts, many predicted to introduce premature termination codons PMID: 24989436
- After a short turnaround time of less than 3 weeks, the diagnosis of CMMR-D could be confirmed by the identification of a homozygous 29-bp deletion in MSH6 (exon 6), which was confirmed by independent methods. PMID: 25431869
- We identified 11 previously unreported mutations in MSH6 in nine different glioma samples and in three oligodendrogliomas and two treatment-naive gliomas PMID: 25078279
- MSH6 founder mutations in Ashkenazi Jews with Lynch syndrome PMID: 23990280
- Some uterine carcinosarcomas show loss of MSH6. PMID: 25083964
- When controlling for surgical and pathologic factors, mismatch repair protein expression did not predict lymph node yield after colectomy for Colon cancer . PMID: 25664706
- The survival time of patients with osteosarcoma may be predicted by local expression of MSH6 and MSH2/6 in surgical primary tumor resections. PMID: 25503122
- isolated loss of expression of hMSH6 in the basal cell layer of oral carcinoma in situ lesions could potentially be a useful diagnostic biomarker for these lesions PMID: 25352643
- Results show that MSH6 and MSH2 expressions are downregulated by YB-1. PMID: 24141788
- DNA mismatch repair gene MSH6 implicated in determining age at natural menopause. PMID: 24357391
- We here demonstrate the causative role of the first germline mutation of MSH2, c.1249-1251 dupGTT (p.417V-418I dupV), associated with normal hMSH2 expression and lack of hMSH6 protein despite a normal MSH6 gene sequence PMID: 25106712
- Immunohistochemistry revealed loss of expression for MLH1, MSH2, MSH6, and PMS2 in 15, 21, 13, and 15 % of cases, respectively...we found a perfect association between MMR immunohistochemical analyses and MSI molecular investigation PMID: 24643686
- This report specifically focus on the protein expression profile and germline mutations of MSH6 and PMS2 genes in 50 Malaysian Lynch syndrome suspected patients. PMID: 24072394
- identified some statistical evidence for an association between the MSH6 G39E polymorphism and risk of cancer PMID: 24622885
- MSH6 protein expression can be a valuable marker to improve prognosis assessment in primary melanoma. PMID: 24926095
- three MSH6 variants found in suspected Lynch syndrome families, MSH6-P1087R, MSH6-R1095H and MSH6-L1354Q were found not to be disease causing when expressed in a mouse model PMID: 24040339
显示更多
收起更多
-
相关疾病:Hereditary non-polyposis colorectal cancer 5 (HNPCC5); Endometrial cancer (ENDMC); Mismatch repair cancer syndrome (MMRCS); Colorectal cancer (CRC)
-
亚细胞定位:Nucleus. Chromosome. Note=Associates with H3K36me3 via its PWWP domain.
-
蛋白家族:DNA mismatch repair MutS family
-
数据库链接:
HGNC: 7329
OMIM: 114500
KEGG: hsa:2956
STRING: 9606.ENSP00000234420
UniGene: Hs.445052
Most popular with customers
-
Recombinant Human Dickkopf-related protein 1 (DKK1) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
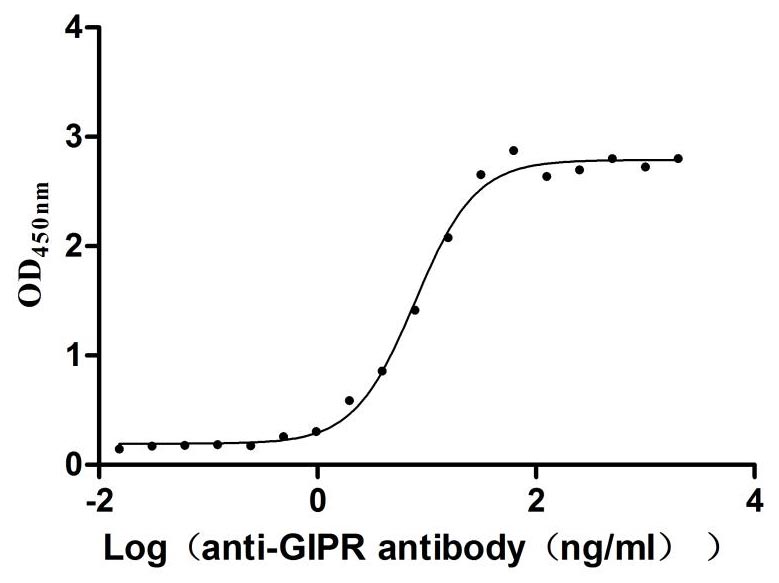
Recombinant Rat Gastric inhibitory polypeptide receptor (Gipr), partial (Active)
Express system: Mammalian cell
Species: Rattus norvegicus (Rat)
-
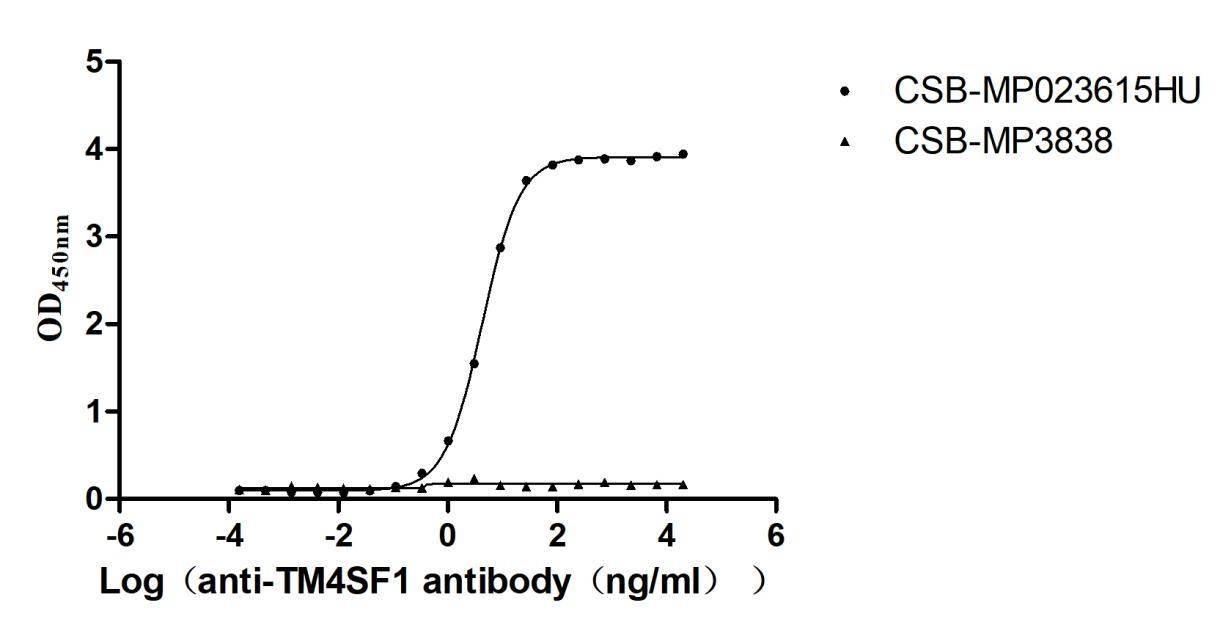
Recombinant Human Transmembrane 4 L6 family member 1(TM4SF1)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human C-C chemokine receptor type 9 (CCR9)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
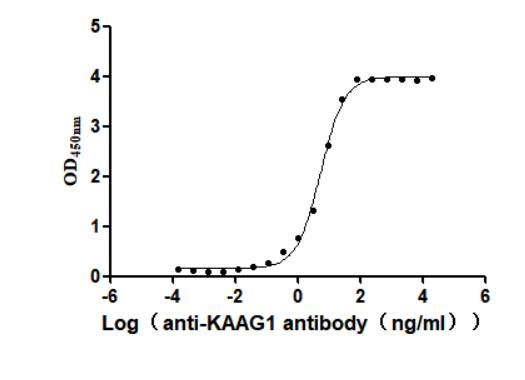
Recombinant Human Kidney-associated antigen 1(KAAG1) (Active)
Express system: Baculovirus
Species: Homo sapiens (Human)


-AC1.jpg)