Recombinant Human Aryl hydrocarbon receptor nuclear translocator-like protein 1 (ARNTL)
-
货号:CSB-YP002123HU
-
规格:
-
来源:Yeast
-
其他:
-
货号:CSB-EP002123HU
-
规格:
-
来源:E.coli
-
其他:
-
货号:CSB-EP002123HU-B
-
规格:
-
来源:E.coli
-
共轭:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
货号:CSB-BP002123HU
-
规格:
-
来源:Baculovirus
-
其他:
-
货号:CSB-MP002123HU
-
规格:
-
来源:Mammalian cell
-
其他:
产品详情
-
纯度:>85% (SDS-PAGE)
-
基因名:ARNTL
-
Uniprot No.:
-
别名:ARNT like protein 1 brain and muscle; Arntl; Aryl hydrocarbon receptor nuclear translocator like; Aryl hydrocarbon receptor nuclear translocator like protein 1; Aryl hydrocarbon receptor nuclear translocator-like protein 1; Basic helix loop helix PAS orphan MOP3; Basic helix loop helix PAS protein MOP3; Basic-helix-loop-helix-PAS protein MOP3; bHLH PAS protein JAP3; bHLH-PAS protein JAP3; bHLHe5; BMAL 1; BMAL1_HUMAN; BMAL1c; Brain and muscle ARNT like 1; Brain and muscle ARNT-like 1; CG8727 PA; Class E basic helix-loop-helix protein 5; cycle; JAP 3; JAP3; Member of PAS protein 3; Member of PAS superfamily 3; MGC47515; MOP 3; MOP3; PAS domain-containing protein 3; PASD 3; PASD3; TIC
-
种属:Homo sapiens (Human)
-
蛋白长度:Full length protein
-
表达区域:1-626
-
氨基酸序列MADQRMDISS TISDFMSPGP TDLLSSSLGT SGVDCNRKRK GSSTDYQESM DTDKDDPHGR LEYTEHQGRI KNAREAHSQI EKRRRDKMNS FIDELASLVP TCNAMSRKLD KLTVLRMAVQ HMKTLRGATN PYTEANYKPT FLSDDELKHL ILRAADGFLF VVGCDRGKIL FVSESVFKIL NYSQNDLIGQ SLFDYLHPKD IAKVKEQLSS SDTAPRERLI DAKTGLPVKT DITPGPSRLC SGARRSFFCR MKCNRPSVKV EDKDFPSTCS KKKADRKSFC TIHSTGYLKS WPPTKMGLDE DNEPDNEGCN LSCLVAIGRL HSHVVPQPVN GEIRVKSMEY VSRHAIDGKF VFVDQRATAI LAYLPQELLG TSCYEYFHQD DIGHLAECHR QVLQTREKIT TNCYKFKIKD GSFITLRSRW FSFMNPWTKE VEYIVSTNTV VLANVLEGGD PTFPQLTASP HSMDSMLPSG EGGPKRTHPT VPGIPGGTRA GAGKIGRMIA EEIMEIHRIR GSSPSSCGSS PLNITSTPPP DASSPGGKKI LNGGTPDIPS SGLLSGQAQE NPGYPYSDSS SILGENPHIG IDMIDNDQGS SSPSNDEAAM AVIMSLLEAD AGLGGPVDFS DLPWPL
-
蛋白标签:Tag type will be determined during the manufacturing process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
产品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
相关产品
靶点详情
-
功能:Transcriptional activator which forms a core component of the circadian clock. The circadian clock, an internal time-keeping system, regulates various physiological processes through the generation of approximately 24 hour circadian rhythms in gene expression, which are translated into rhythms in metabolism and behavior. It is derived from the Latin roots 'circa' (about) and 'diem' (day) and acts as an important regulator of a wide array of physiological functions including metabolism, sleep, body temperature, blood pressure, endocrine, immune, cardiovascular, and renal function. Consists of two major components: the central clock, residing in the suprachiasmatic nucleus (SCN) of the brain, and the peripheral clocks that are present in nearly every tissue and organ system. Both the central and peripheral clocks can be reset by environmental cues, also known as Zeitgebers (German for 'timegivers'). The predominant Zeitgeber for the central clock is light, which is sensed by retina and signals directly to the SCN. The central clock entrains the peripheral clocks through neuronal and hormonal signals, body temperature and feeding-related cues, aligning all clocks with the external light/dark cycle. Circadian rhythms allow an organism to achieve temporal homeostasis with its environment at the molecular level by regulating gene expression to create a peak of protein expression once every 24 hours to control when a particular physiological process is most active with respect to the solar day. Transcription and translation of core clock components (CLOCK, NPAS2, ARNTL/BMAL1, ARNTL2/BMAL2, PER1, PER2, PER3, CRY1 and CRY2) plays a critical role in rhythm generation, whereas delays imposed by post-translational modifications (PTMs) are important for determining the period (tau) of the rhythms (tau refers to the period of a rhythm and is the length, in time, of one complete cycle). A diurnal rhythm is synchronized with the day/night cycle, while the ultradian and infradian rhythms have a period shorter and longer than 24 hours, respectively. Disruptions in the circadian rhythms contribute to the pathology of cardiovascular diseases, cancer, metabolic syndromes and aging. A transcription/translation feedback loop (TTFL) forms the core of the molecular circadian clock mechanism. Transcription factors, CLOCK or NPAS2 and ARNTL/BMAL1 or ARNTL2/BMAL2, form the positive limb of the feedback loop, act in the form of a heterodimer and activate the transcription of core clock genes and clock-controlled genes (involved in key metabolic processes), harboring E-box elements (5'-CACGTG-3') within their promoters. The core clock genes: PER1/2/3 and CRY1/2 which are transcriptional repressors form the negative limb of the feedback loop and interact with the CLOCK|NPAS2-ARNTL/BMAL1|ARNTL2/BMAL2 heterodimer inhibiting its activity and thereby negatively regulating their own expression. This heterodimer also activates nuclear receptors NR1D1/2 and RORA/B/G, which form a second feedback loop and which activate and repress ARNTL/BMAL1 transcription, respectively. ARNTL/BMAL1 positively regulates myogenesis and negatively regulates adipogenesis via the transcriptional control of the genes of the canonical Wnt signaling pathway. Plays a role in normal pancreatic beta-cell function; regulates glucose-stimulated insulin secretion via the regulation of antioxidant genes NFE2L2/NRF2 and its targets SESN2, PRDX3, CCLC and CCLM. Negatively regulates the mTORC1 signaling pathway; regulates the expression of MTOR and DEPTOR. Controls diurnal oscillations of Ly6C inflammatory monocytes; rhythmic recruitment of the PRC2 complex imparts diurnal variation to chemokine expression that is necessary to sustain Ly6C monocyte rhythms. Regulates the expression of HSD3B2, STAR, PTGS2, CYP11A1, CYP19A1 and LHCGR in the ovary and also the genes involved in hair growth. Plays an important role in adult hippocampal neurogenesis by regulating the timely entry of neural stem/progenitor cells (NSPCs) into the cell cycle and the number of cell divisions that take place prior to cell-cycle exit. Regulates the circadian expression of CIART and KLF11. The CLOCK-ARNTL/BMAL1 heterodimer regulates the circadian expression of SERPINE1/PAI1, VWF, B3, CCRN4L/NOC, NAMPT, DBP, MYOD1, PPARGC1A, PPARGC1B, SIRT1, GYS2, F7, NGFR, GNRHR, BHLHE40/DEC1, ATF4, MTA1, KLF10 and also genes implicated in glucose and lipid metabolism. Promotes rhythmic chromatin opening, regulating the DNA accessibility of other transcription factors. The NPAS2-ARNTL/BMAL1 heterodimer positively regulates the expression of MAOA, F7 and LDHA and modulates the circadian rhythm of daytime contrast sensitivity by regulating the rhythmic expression of adenylate cyclase type 1 (ADCY1) in the retina. The preferred binding motif for the CLOCK-ARNTL/BMAL1 heterodimer is 5'-CACGTGA-3', which contains a flanking Ala residue in addition to the canonical 6-nucleotide E-box sequence. CLOCK specifically binds to the half-site 5'-CAC-3', while ARNTL binds to the half-site 5'-GTGA-3'. The CLOCK-ARNTL/BMAL1 heterodimer also recognizes the non-canonical E-box motifs 5'-AACGTGA-3' and 5'-CATGTGA-3'. Essential for the rhythmic interaction of CLOCK with ASS1 and plays a critical role in positively regulating CLOCK-mediated acetylation of ASS1. Plays a role in protecting against lethal sepsis by limiting the expression of immune checkpoint protein CD274 in macrophages in a PKM2-dependent manner. Regulates the diurnal rhythms of skeletal muscle metabolism via transcriptional activation of genes promoting triglyceride synthesis (DGAT2) and metabolic efficiency (COQ10B).
-
基因功能参考文献:
- BMAL1 Deficiency Contributes to Mandibular Dysplasia by Upregulating MMP3. PMID: 29276151
- ARNTL rs7107287 was associated with a cyclothymic temperament, depressive and stress symptoms PMID: 28708003
- NR1D1 and BMAL1 mRNA and protein levels were significantly reduced in OA compared to normal cartilage. In cultured human chondrocytes, a clear circadian rhythmicity was observed for NR1D1 and BMAL1. PMID: 27884645
- These results suggest that the circadian clock system can be recovered through BMAL1 expression induced by aza-dC within a day. PMID: 28487473
- Determined a novel role of TNF-alpha in inducing Bmal1 via dual calcium dependent pathways; Roralpha was up-regulated in the presence of Ca(2+) influx and Rev-erbalpha was down-regulated in the absence of that. PMID: 29217191
- results of this study suggest that genetic variability in the ARNTL and CLOCK genes might be associated with risk for multiple sclerosis PMID: 29324865
- PI3K-PTEN upregulated-mTORC1 and mTORC2 complex plays a critical role in controlling BMAL1, establishing a connection between PI3K signaling and the regulation of circadian rhythm, ultimately resulting in deregulated BMAL1 in tumor cells with disrupted PI3K signaling PMID: 27285754
- M. tuberculosis infection caused enhanced MMP-1, -9, and miR-223 expression, with inhibited BMAL1 expression. MiR-223 modulated BMAL1 expression via the direct binding of BMAL1 3'-UTR. PMID: 28543681
- research describes an association between changes in the methylation of the BMAL1 gene with the intervention and the effects of a weight loss intervention on blood lipids levels PMID: 26873744
- Study found rhythmic methylation of BMAL1 was altered in Alzheimer's disease brains and fibroblasts and correlated with transcription cycles. Results indicate that cycles of DNA methylation contribute to the regulation of BMAL1 rhythms in the brain. PMID: 27883893
- TFEB regulates PER3 expression via glucose-dependent effects on CLOCK/BMAL1 PMID: 27373683
- found that overexpression of both Clock and Bmal1 suppressed cell growth PMID: 26370682
- our results identified Bmal1 as a novel tumor suppressor gene that elevates the sensitivity of cancer cells to paclitaxel, with potential implications as a chronotherapy timing biomarker in tongue squamous cell carcinoma PMID: 27821487
- The level of BMAL1 expression in granulosa cells the polycystic ovary syndrome (PCOS) group was lower than that of the group without PCOS. We also analyzed estrogen synthesis and aromatase expression in KGN cell lines. Both were downregulated after BMAL1 and SIRT1 knock-down and, conversely, upregulated after overexpression treatments of these two genes in KGN cells. PMID: 27117143
- this study shows that BMAL1 can regulate cellular innate immunity against specific RNA viruses PMID: 27913791
- Bmal1 is a key clock gene to involve in cartilage homeostasis mediated through sirt1. PMID: 27253997
- The BMAL1 rs2278749 T/C was associated with Alzheimer disease (AD) risk, and T carriers in BMAL1 rs2278749 T/C showed a higher risk of AD than did non-carriers. PMID: 26782499
- Our results indicate that activation of TGF-beta1 promotes the transcriptional induction of BMAL1. PMID: 26753996
- possible circadian rhythm in full-term placental expression PMID: 26247999
- Synchronized cells exhibit an autonomous ultradian mitochondrial respiratory activity which is abrogated by silencing the master clock gene BMAL1. PMID: 27060253
- decreased expression of Bmal1 is correlated with tumor progression and poor prognosis in pancreatic ductal adenocarcinoma, with potential to be used as a biomarker for diagnosis and prognosis PMID: 26915801
- ARNTL and PER1 were associated with PD. PMID: 26507264
- define a regulatory mechanism that links chondrocyte BMAL1 to the maintenance and repair of cartilage PMID: 26657859
- when overexpressed, c-MYC is able to repress Per1 transactivation by BMAL1/CLOCK via targeting selective E-box sequences. Importantly, upon serum stimulation, MYC was detected in BMAL1 protein complexes PMID: 26850841
- Bmal1 could directly bind to the p53 gene promoter and thereby transcriptionally activate the downstream tumor suppressor pathway in a p53-dependent manner in pancreatic tumors. PMID: 26683776
- Data suggest that cryptochromes mediate periodic binding of Ck2b (casein kinase 2beta) to Bmal1 (aryl hydrocarbon receptor nuclear translocator-like protein) and thus inhibit Bmal1-Ser90 phosphorylation by Ck2a (casein kinase 2alpha). [SYNOPSIS] PMID: 26562283
- a 4-locus CSNK1E haplotype encompassing the rs1534891 SNP (Z-score=2.685, permuted p=0.0076) and a 3-locus haplotype in ARNTL (Z-score=3.269, permuted p=0.0011) showed a significant association with Bipolar Disorder PMID: 26283580
- CLOCK, ARNTL, and NPAS2 gene polymorphisms may have a role in seasonal variations in mood and behavior PMID: 26134245
- rs2290036-C variant of ARNTL was over-represented in psychosis patients, and the variants rs934945-G and rs10462023-G of PER2 were associated with a more severe psychotic disorder PMID: 25799324
- these findings suggest that sumoylation plays a critical role in the spatiotemporal co-activation of CLOCK-BMAL1 by CBP for immediate-early Per induction and the resetting of the circadian clock. PMID: 26164627
- In men undergoing acute total sleep deprivation, BMAL1 gene expression was decreased in skeletal muscle compared with controls. PMID: 26168277
- Data indicate that the inner nuclear membrane protein MAN1 directly binds the transcription activator BMAL1 promoter and enhances its transcription. PMID: 25182847
- Bmal1-dependent oscillators of arginine vasopressin neurons modulate the coupling of the suprachiasmatic nucleus. PMID: 25741730
- PER1 and BMAL1 operate as cell-autonomous modulators of human pigmentation and may be targeted for future therapeutic strategies PMID: 25310406
- These results suggested that ARNTL may be a tumor suppressor and is epigenetically silenced in ovarian cancer. PMID: 25175925
- temporal signals of fasting and refeeding hormones regulate the transcription of Bmal1, a key transcription activator of molecular clock, in the liver PMID: 25480789
- There is not a significant difference in the expression of CLOCK, BMAL1, and PER1 in buccal epithelial cells of patients with essential arterial hypertension regardless of patient genotype. PMID: 25070164
- FAL1 associates with the epigenetic repressor BMI1 and regulates its stability in order to modulate the transcription of a number of genes including CDKN1A. PMID: 25203321
- The results of this study suggest that the ARNTL gene may be associated with the lithium prophylactic response in bipolar illness. PMID: 24636202
- The SNPs most strongly associated in the single-marker analysis of the combined Danish samples were rs4757144 in ARNTL (P=3.78 x 10-6) and rs8057927 in CDH13. PMID: 23358160
- The progression-free survival of patients with high Bmal1 expression is significantly longer than that of patients with low Bmal1 expression. PMID: 24277452
- The rhythm of Bmal1 mRNA in human plaque-derived vascular smooth muscle cells is altered. PMID: 24418196
- These results suggest that DNA methylation of the BMAL1 gene is critical for interfering with circadian rhythms. PMID: 24103761
- Knockdown of either BMAL1 or Period1 in human anagen hair follicles significantly prolonged anagen. PMID: 24005054
- there is significant daily variation in PER2, PER3, and ARNTL1 expression with earlier timing of expression in women than in men PMID: 23606611
- Bmal1 is a tumor suppressor, capable of suppressing cancer cell growth and invasiveness. PMID: 23563360
- The data suggest that the impairment of the BMAL1 clock gene expression is closely associated with GDM susceptibility. PMID: 23206673
- O-GlcNAc transferase (OGT) promotes expression of BMAL1/CLOCK target genes and affects circadian oscillation of clock genes in vitro and in vivo. PMID: 23395176
- Variants in ARNTL have been associated with seasonality and seasonal affective disorder, phenotypes that could reflect circadian rhythm disruption. PMID: 23449886
- The phospho-mimicking S78E mutant of BMAL1 efficiently blocks DNA binding, which provides a molecular rationale for the possibility of rhythmic binding of CLOCK-BMAL1 during circadian cycle. PMID: 23229515
显示更多
收起更多
-
亚细胞定位:Nucleus. Cytoplasm. Nucleus, PML body.
-
组织特异性:Hair follicles (at protein level). Highly expressed in the adult brain, skeletal muscle and heart.
-
数据库链接:
HGNC: 701
OMIM: 602550
KEGG: hsa:406
STRING: 9606.ENSP00000374357
UniGene: Hs.65734
Most popular with customers
-
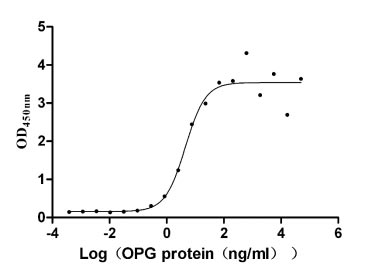
Recombinant Human Tumor necrosis factor receptor superfamily member 11B (TNFRSF11B) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
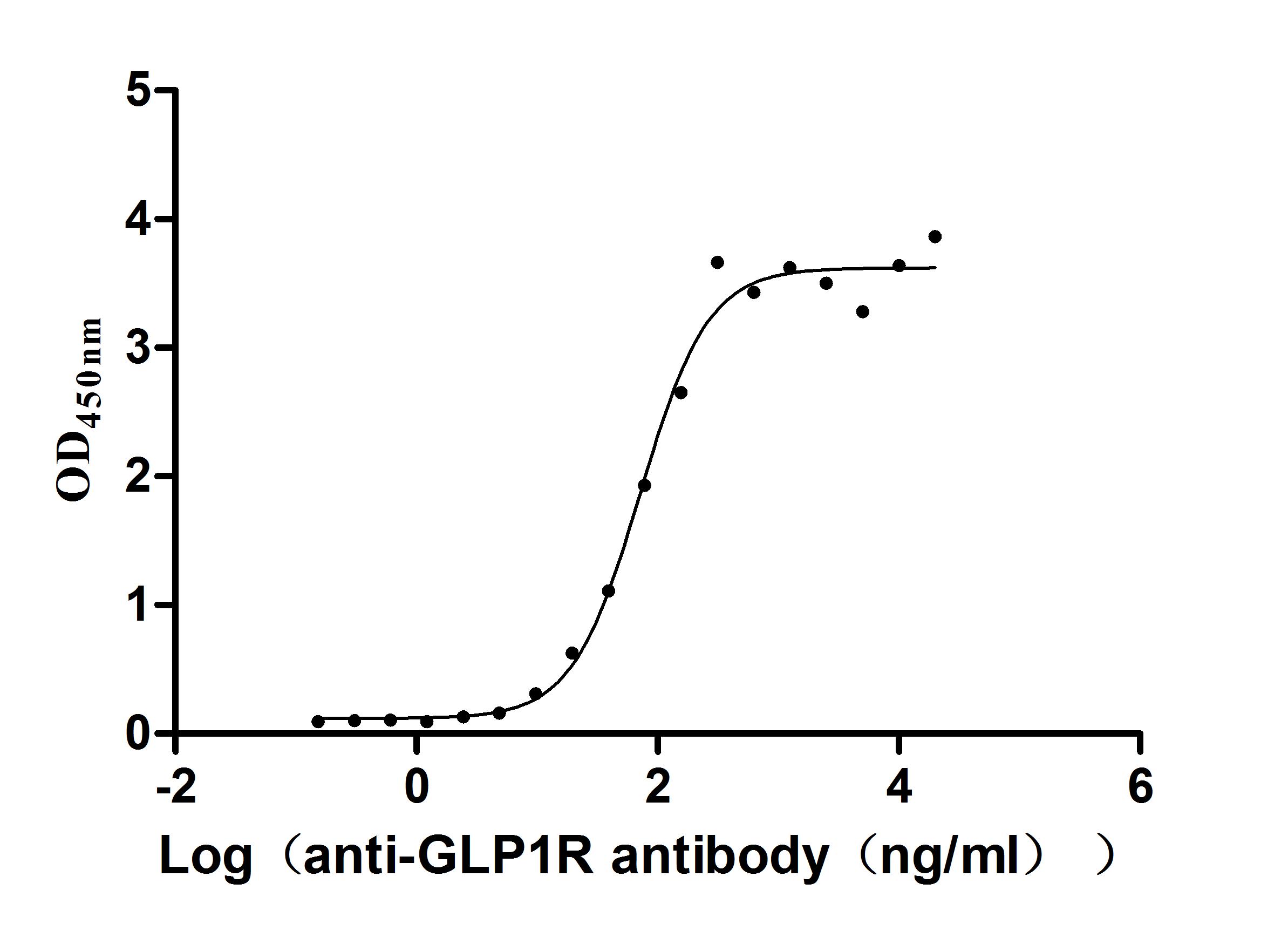
Recombinant Human Glucagon-like peptide 1 receptor (GLP1R), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
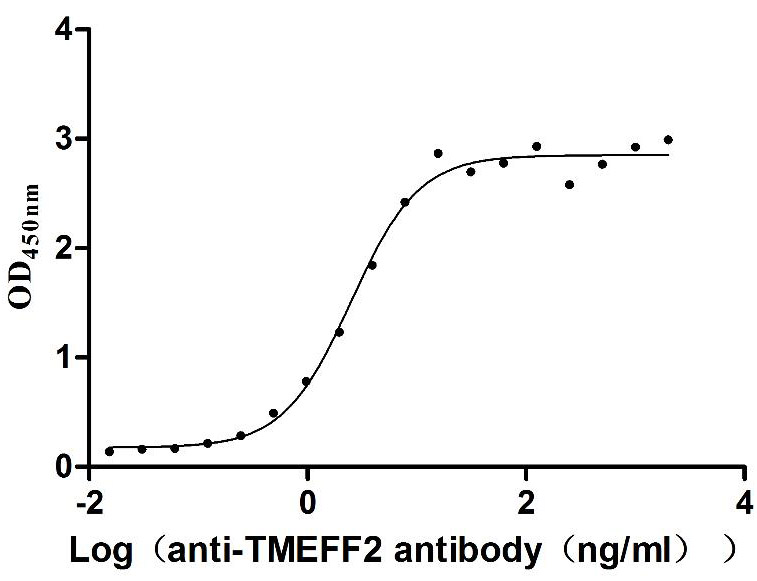
Recombinant Human Tomoregulin-2 (TMEFF2), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human C-type lectin domain family 4 member C (CLEC4C), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human C-C chemokine receptor type 6(CCR6)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Macaca fascicularis Cadherin 6(CDH6),partial (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
Recombinant Human Tumor necrosis factor ligand superfamily member 15(TNFSF15) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
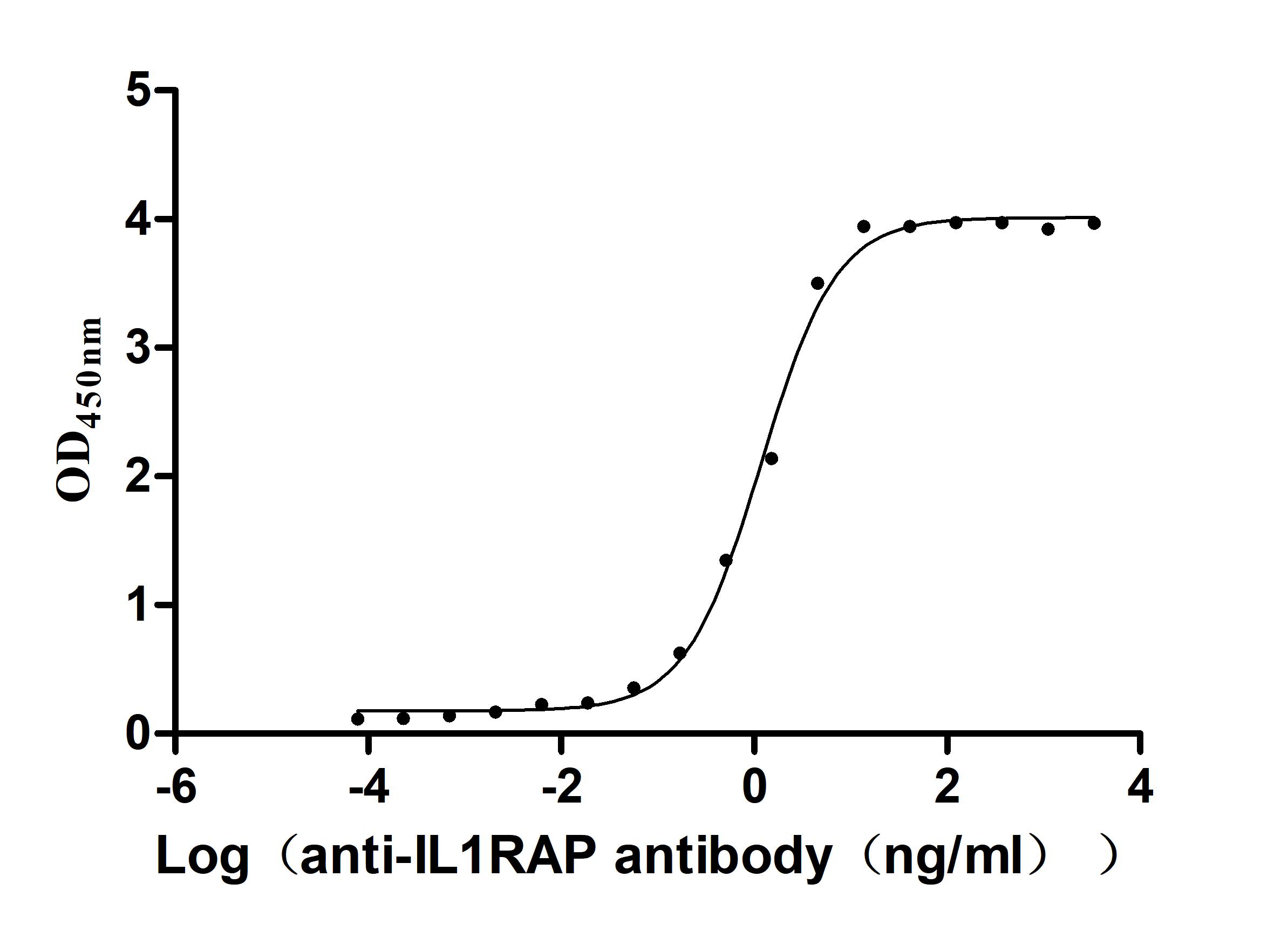
Recombinant Human Interleukin-1 receptor accessory protein (IL1RAP), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)


-AC1.jpg)