Human Histone RNA hairpin-binding protein(SLBP) ELISA kit
-
中文名称:人组蛋白RNA发夹结合蛋白(SLBP)酶联免疫试剂盒
-
货号:CSB-EL021372HU
-
规格:96T/48T
-
价格:¥3600/¥2500
-
其他:
产品详情
-
产品描述:
This Human SLBP ELISA Kit was designed for the quantitative measurement of Human SLBP protein in serum, plasma, tissue homogenates, cell lysates. It is a Sandwich ELISA kit, its detection range is 15.6 pg/mL-1000 pg/mL and the sensitivity is 3.9 pg/mL.
-
别名:Hairpin binding protein histone ELISA Kit; HBP ELISA Kit; Histone binding protein ELISA Kit; Histone RNA hairpin binding protein ELISA Kit; Histone RNA hairpin-binding protein ELISA Kit; Histone stem-loop-binding protein ELISA Kit; SLBP ELISA Kit; SLBP_HUMAN ELISA Kit; Stem loop (histone) binding protein ELISA Kit; Stem loop binding protein ELISA Kit
-
缩写:SLBP
-
Uniprot No.:
-
种属:Homo sapiens (Human)
-
样本类型:serum, plasma, tissue homogenates, cell lysates
-
检测范围:15.6 pg/mL-1000 pg/mL
-
灵敏度:3.9 pg/mL
-
反应时间:1-5h
-
样本体积:50-100ul
-
检测波长:450 nm
-
研究领域:Epigenetics and Nuclear Signaling
-
测定原理:quantitative
-
测定方法:Sandwich
-
精密度:
Intra-assay Precision (Precision within an assay): CV%<8% Three samples of known concentration were tested twenty times on one plate to assess. Inter-assay Precision (Precision between assays): CV%<10% Three samples of known concentration were tested in twenty assays to assess. -
线性度:
To assess the linearity of the assay, samples were spiked with high concentrations of human SLBP in various matrices and diluted with the Sample Diluent to produce samples with values within the dynamic range of the assay. Sample Serum(n=4) 1:1 Average % 91 Range % 86-95 1:2 Average % 102 Range % 97-107 1:4 Average % 91 Range % 85-97 1:8 Average % 97 Range % 91-103 -
回收率:
The recovery of human SLBP spiked to levels throughout the range of the assay in various matrices was evaluated. Samples were diluted prior to assay as directed in the Sample Preparation section. Sample Type Average % Recovery Range Serum (n=5) 95 89-98 EDTA plasma (n=4) 97 90-100 -
标准曲线:
These standard curves are provided for demonstration only. A standard curve should be generated for each set of samples assayed. 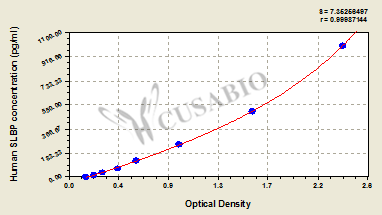
pg/ml OD1 OD2 Average Corrected 1000 2.338 2.398 2.368 2.206 500 1.587 1.599 1.593 1.431 250 0.949 0.968 0.959 0.797 125 0.599 0.589 0.594 0.432 62.5 0.425 0.436 0.431 0.269 31.25 0.293 0.316 0.305 0.143 15.6 0.231 0.222 0.227 0.065 0 0.161 0.163 0.162 -
数据处理:
-
货期:3-5 working days
相关产品
靶点详情
-
功能:RNA-binding protein involved in the histone pre-mRNA processing. Binds the stem-loop structure of replication-dependent histone pre-mRNAs and contributes to efficient 3'-end processing by stabilizing the complex between histone pre-mRNA and U7 small nuclear ribonucleoprotein (snRNP), via the histone downstream element (HDE). Plays an important role in targeting mature histone mRNA from the nucleus to the cytoplasm and to the translation machinery. Stabilizes mature histone mRNA and could be involved in cell-cycle regulation of histone gene expression. Involved in the mechanism by which growing oocytes accumulate histone proteins that support early embryogenesis. Binds to the 5' side of the stem-loop structure of histone pre-mRNAs.
-
基因功能参考文献:
- SLBP is a potentially important cellular regulator of HIV-1, thereby establishing a link between histone metabolism, inflammation, and HIV-1 infection PMID: 27454292
- showed that inhibiting the SLBP mRNA and protein levels were rescued by epigenetic modifiers suggesting that nickel's effects on SLBP may be mediated via epigenetic mechanisms PMID: 28306745
- Cyclin F-mediated degradation of SLBP limits H2A.X accumulation and apoptosis upon genotoxic stress in G2 cell cycle checkpoint. PMID: 27773672
- CRL4(WDR23) is required for efficient histone mRNA 3' end processing to produce mature histone mRNAs for translation. CRL4(WDR23) binds and ubiquitylates SLBP in vitro and in vivo, and this modification activates SLBP function in histone mRNA 3' end processing without affecting its protein levels. PMID: 27203182
- FEM1 proteins are ancient regulators of Stem-Loop Binding Protein. PMID: 28118078
- CRL4-DCAF11 mediates the degradation of SLBP at the end of S phase and this degradation is essential for the viability of cells. PMID: 27254819
- arsenic, a carcinogenic metal, decreases cellular levels of SLBP by inducing its proteasomal degradation and inhibiting SLBP transcription via epigenetic mechanisms PMID: 25266719
- the S/G2 stable mutant form of SLBP is degraded by proteasome in G1, indicating that indicating that the SLBP degradation in G1 is independent of the previously identified SLBP degradation at S/G2 PMID: 24122909
- Although the C-terminal tail of dSLBP does not contact the RNA, phosphorylation of the tail promotes SLBP conformations competent for RNA binding and thereby appears to reduce the entropic penalty for the association. PMID: 25002523
- Alternative splicing allows the synthesis of HBP/SLBP isoforms with different properties that may be important for regulating HBP/SLBP functions during replication stress. PMID: 23941746
- C-terminal extension of Lsm4 interacts directly with the histone mRNP, contacting both SLBP and 3'hExo. PMID: 24255165
- Data suggest that oligomerization and SLBP phosphorylation regulate SLBP-SLIP1-histone-mRNA complex formation/disassociation; sequential and ordered assembly is required. PMID: 23286197
- Using yeast two-hybrid screening, the s identify CT initiation factor-interacting protein (CTIF) as a protein that binds directly to SLBP. SLBP preferentially associates with the CT complex of histone mRNAs consisting of CBP80/CBP20, but not with the eIF4E/eIF4G (ET) complex, as has been proposed. Rapid degradation of histone mRNA on the inhibition of DNA replication requires association of SLBP with CTIF. PMID: 23234701
- This paper shows that SLBP is a substrate for the prolyl isomerase Pin1. Pin1, along with PP2A, facilitates dissociation of the SLBP-histone mRNA complex at the end of S-phase, thereby promoting histone mRNA decay and SLBP ubiquitination. PMID: 22907757
- This paper describes the biophysical characterization of human SLBP and the SLBP-SLIP1 complex. Human SLBP is an intrinsically disordered protein that is phosphorylated at 23 Ser/Thr sites when expressed in a eukaryotic expression system such as baculovirus. Unphosphorylated human SLBP forms a high affinity heterotetramer with SLIP1 and the SLBP-SLIP1 complex is regulated by SLBP phosphorylation. PMID: 23286197
- the crystal structure of a ternary complex of human SLBP RNA binding domain, human 3'hExo, and a 26-nucleotide stem-loop RNA is reported. PMID: 23329046
- The nuclear magnetic resonance and kinetic studies presented here provide a framework for understanding how SLBP recognizes histone mRNA and highlight possible structural roles of phosphorylation and proline isomerization in RNA binding proteins PMID: 22439849
- SLBP is the only cell cycle-regulated factor required for histone pre-mRNA processing PMID: 12588979
- human HBP/SLBP is essential for the coordinate synthesis of DNA and histone proteins and is required for progression through the cell division cycle PMID: 15546920
- SLBP is imported into the cell nucleus during the cell cycle. PMID: 15829567
- SLBP is required for efficient DNA replication probably because a decreased ability to assemble chromatin results in a decrease in the rate of DNA replication PMID: 15916543
- Removal of the phosphoryl group from T230 by either dephosphorylation or mutation results in a 7-fold reduction in the affinity of SLBP for the stem-loop RNA PMID: 16492733
- replication-dependent histone mRNAs are likely to be the sole target of SLBP PMID: 16931877
- SLBP is required for efficient histone 3'-UTR end processing. PMID: 16982637
- Here we show that NELF interacts with the cap binding complex (CBC), a factor that plays important roles in mRNA processing steps, and the two factors together participate in the 3' end processing of histone mRNAs, through association with the SLBP. PMID: 17499042
- identified five conserved residues in a 15-amino-acid region in the amino-terminal portion of SLBP, each of which is required for translation. PMID: 18025107
- Study concludes that the increase in cyclin A/Cdk1 activity at the end of S phase triggers degradation of SLBP at S/G(2). PMID: 18490441
- These results suggest a previously undescribed role for SLBP in histone mRNA export. PMID: 19155325
显示更多
收起更多
-
亚细胞定位:Cytoplasm. Nucleus.
-
蛋白家族:SLBP family
-
组织特异性:Widely expressed.
-
数据库链接:
Most popular with customers
-
Human Transforming Growth factor β1,TGF-β1 ELISA kit
Detect Range: 23.5 pg/ml-1500 pg/ml
Sensitivity: 5.8 pg/ml
-
-
-
Mouse Tumor necrosis factor α,TNF-α ELISA Kit
Detect Range: 7.8 pg/ml-500 pg/ml
Sensitivity: 1.95 pg/ml
-
-
-
-













