Recombinant Arabidopsis thaliana MADS-box protein FLOWERING LOCUS C (FLC)
-
中文名称:拟南芥FLC重组蛋白
-
货号:CSB-YP890276DOA
-
规格:
-
来源:Yeast
-
其他:
-
中文名称:拟南芥FLC重组蛋白
-
货号:CSB-EP890276DOA
-
规格:
-
来源:E.coli
-
其他:
-
中文名称:拟南芥FLC重组蛋白
-
货号:CSB-EP890276DOA-B
-
规格:
-
来源:E.coli
-
共轭:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
中文名称:拟南芥FLC重组蛋白
-
货号:CSB-BP890276DOA
-
规格:
-
来源:Baculovirus
-
其他:
-
中文名称:拟南芥FLC重组蛋白
-
货号:CSB-MP890276DOA
-
规格:
-
来源:Mammalian cell
-
其他:
产品详情
-
纯度:>85% (SDS-PAGE)
-
基因名:FLC
-
Uniprot No.:
-
别名:FLC; FLF; At5g10140; T31P16.130MADS-box protein FLOWERING LOCUS C; MADS-box protein FLOWERING LOCUS F
-
种属:Arabidopsis thaliana (Mouse-ear cress)
-
蛋白长度:full length protein
-
表达区域:1-196
-
氨基酸序列MGRKKLEIKR IENKSSRQVT FSKRRNGLIE KARQLSVLCD ASVALLVVSA SGKLYSFSSG DNLVKILDRY GKQHADDLKA LDHQSKALNY GSHYELLELV DSKLVGSNVK NVSIDALVQL EEHLETALSV TRAKKTELML KLVENLKEKE KMLKEENQVL ASQMENNHHV GAEAEMEMSP AGQISDNLPV TLPLLN
-
蛋白标签:Tag type will be determined during the manufacturing process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
产品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
靶点详情
-
功能:Putative transcription factor that seems to play a central role in the regulation of flowering time in the late-flowering phenotype by interacting with 'FRIGIDA', the autonomous and the vernalization flowering pathways. Inhibits flowering by repressing 'SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1'.
-
基因功能参考文献:
- FLC cis elements are recognized by VAL1 specific B3 domain during vernalization. PMID: 29733847
- PWWP domain proteins function together with FVE and MSI5 to regulate the function of the PRC2 complex on FLC. PMID: 29314758
- For seed dormancy, FT regulates seed dormancy through FLC gene expression and regulates chromatin state by activating antisense FLC transcription. PMID: 29853684
- The results suggest that TAF15b affects flowering time through transcriptional repression of FLC in Arabidopsis. PMID: 29086456
- FLC silencing is inherited through in vitro regeneration PMID: 28498984
- The vernalization insensitivity caused by mutant COLDAIR was rescued by the ectopic expression of the wild-type COLDAIR. Our study reveals the molecular framework in which COLDAIR lncRNA mediates the PRC2-mediated repression of FLC during vernalization. PMID: 28759577
- The relationship between H2AK121ub and H3K27me3 marks across the A. thaliana genome and unveil that ubiquitination by PRC1 is largely independent of PRC2 activity in plants, while the inverse is true for H3K27 trimethylation. PMID: 28403905
- this study identified a novel bona fide SUMO protease, ASP1, which positively regulates transition to flowering at least partly by repressing FLC protein stability. PMID: 27925396
- these findings report that the interaction between MADS box transcription factor FLC and GRAS domain regulator DELLAs may integrate various signaling inputs in flowering time control, and shed new light on the regulatory mechanism both for FLC and DELLAs in regulating gene expression. PMID: 26584710
- genotypes, many of which have high levels of the floral repressor FLOWERING LOCUS C (FLC), that bolted dramatically earlier in fluctuating - as opposed to constant - warm temperatures, were identified. PMID: 26681345
- SKIP interacted with the Paf1c to modulate the expression of FLC clade genes at the transcriptional level. PMID: 26384244
- FLC appears as a major modulator of the natural variation for the plasticity of flowering to multiple environmental factors. PMID: 26173848
- propose that BRR2a is specifically needed for efficient splicing of a subset of introns characterized by a combination of factors including intron size, sequence and chromatin, and that FLC is most sensitive to splicing defects PMID: 27100965
- INDUCER OF CBF EXPRESSION 1 integrates cold signals into FLOWERING LOCUS C-mediated flowering pathways in Arabidopsis PMID: 26248809
- Intragenic methylation triggered by RNA-directed DNA methylation (RdDM) promoted FT expression. DNA methylation of the FT gene body blocked flowering locus C (FLC)repressor binding to the CArG boxes. PMID: 26076969
- this study investigated how FCA and FLD transcriptionally repress FLC through analysis of Pol II occupancy. PMID: 26699513
- Epigenetic memory of FLC expression is stored not in trans memory but in cis memory. PMID: 25955967
- cold temperature exposure is likely to be registered in an all-or-nothing (digital) manner at the relevant gene FLOWERING LOCUS C PMID: 25775579
- a single natural intronic polymorphism in one haplotype affects FLC expression and thus flowering by specifically changing splicing of the FLC antisense transcript COOLAIR. PMID: 25805848
- genetic analysis showed COOLAIR and Polycomb complexes work independently in the cold-dependent silencing of FLC. PMID: 25349421
- For many phases of the vernalization process H3K36me3 and H3K27me3 show opposing profiles in the FLC nucleation region and gene body, and H3K36me3 and H3K27me3 rarely coexist on the same histone tail; this antagonism is functionally important. PMID: 25065750
- The expression of FLC gene in Arabidopsis thaliana plants in extreme conditions in northern margins of species range. PMID: 25474881
- BAF60 creates a repressive chromatin configuration at the FLC locus. PMID: 24510722
- five predominant FLC haplotypes defined by noncoding sequence variation. Genetic and transgenic experiments show that they are functionally distinct, varying in FLC expression level and rate of epigenetic silencing PMID: 25035417
- Sumoylation of FLC is critical for its role in the control of flowering time and that AtSIZ1 positively regulates FLC-mediated floral suppression. PMID: 24218331
- UGT87A2 regulates flowering time via the flowering repressor FLOWERING LOCUS C. PMID: 22404750
- cdkc;2 specifically reduces transcription of COOLAIR antisense transcripts, which indirectly up-regulates FLC expression through disruption of a COOLAIR-mediated repression mechanism. PMID: 24799695
- Suggest that altered splicing of a long noncoding transcript COOLAIR can quantitatively modulate FLC gene expression through cotranscriptional coupling mechanisms. PMID: 24725596
- FLC protein plays role in vernalization and de-vernalization responses. PMID: 23581257
- the dynamics of FLC transcription and associated histone H3K27me3 activity are closely linked biologically PMID: 22543923
- FLC gene family are differentially regulated during the course of vernalization to mediate proper vernalization response. PMID: 23417034
- The FLOWERING LOCUS C clade members act as part of several MADS-domain complexes with partial redundancy, which integrate responses to endogenous and environmental cues to control flowering. PMID: 23770815
- The mutations of AtPRMT10 derepress FLOWERING LOCUS C (FLC) expression resulting in a late-flowering phenotype. PMID: 22729397
- These results are consistent with Del(-57) allele acting as a novel cis-regulatory FLC polymorphism that may confer climatic adaptation by increasing vernalization sensitivity. PMID: 22494398
- The FLC loop is disrupted during vernalization, the cold-induced, Polycomb-dependent epigenetic silencing of FLC. Loop disruption parallels timing of the cold-induced FLC transcriptional shut-down and upregulation of FLC antisense transcripts. PMID: 23222483
- vacuolar and/or endocytic trafficking is involved in the FLC regulation of flowering time in A. thaliana PMID: 22848750
- Current understanding of the molecular mechanism of vernalization-mediated FLC silencing, is described. PMID: 22078062
- study concludes that DCL4 promotes transcription termination of the FCA gene, reducing the amount of aberrant RNA produced from the locus PMID: 22461611
- MSI5 acts in partial redundancy with FVE to silence FLOWERING LOCUS C (FLC), which is a crucial floral repressor subject to asRNA-mediated silencing, FLC homologs PMID: 22102827
- Data show that the promoter and first exon of the FLC gene are sufficient to initiate repression during vernalization; this initial repression of FLC does not require antisense transcription. PMID: 21713009
- effect of changes in transcription rate on the abundance of H3K27me3 in the FLC gene body, a chromatin region that includes sequences required to maintain FLC repression following vernalization PMID: 21276103
- Formation of the FRI complex leads to the active chromatin state of the FLC gene. PMID: 21282526
- These results indicate novel and FCA-independent roles for FY in the regulation of FLC. PMID: 21209277
- The transcriptional activation of FLC, how different activities are integrated at this one locus and why FLC regulation seems so sensitive to mutation in these conserved gene regulatory pathways, are discussed. PMID: 20884277
- results demonstrate that the onset and the progression of vegetative phase change are regulated by different combinations of endogenous and environmental factors, and reveal a role for FLC in vegetative development PMID: 21228003
- findings show that a long intronic noncoding RNA (COLDAIR)] is required for the vernalization-mediated epigenetic repression of FLC; COLDAIR physically associates with a component of PRC2 and targets PRC2 to FLC PMID: 21127216
- CDC73 is required for high levels of FLC expression in a subset of autonomous-pathway-mutant backgrounds and functions both to promote activating histone modifications (H3K4me3) as well as preventing repressive ones (e.g. H3K27me3). PMID: 20463090
- analysis of control of seasonal expression of the Arabidopsis FLC gene in a fluctuating environment PMID: 20534541
- Both classes of H3K4 methylases, atx1 and atxr7, appear to be required for proper regulation of FLC expression. PMID: 19855050
- Data suggest that FLC is repressed via a novel pathway involving the SIR2 class of histone deacetylases. PMID: 19825652
显示更多
收起更多
-
亚细胞定位:Nucleus.
-
组织特异性:High expression in the vegetative apex and in root tissue and lower expression in leaves and stems. Not detected in young tissues of the inflorescence. Before fertilization, expressed in ovules, but not in pollen or stamens, of non-vernalized plants. Afte
-
数据库链接:
KEGG: ath:AT5G10140
STRING: 3702.AT5G10140.1
UniGene: At.1629
Most popular with customers
-
Recombinant Human Dickkopf-related protein 1 (DKK1) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
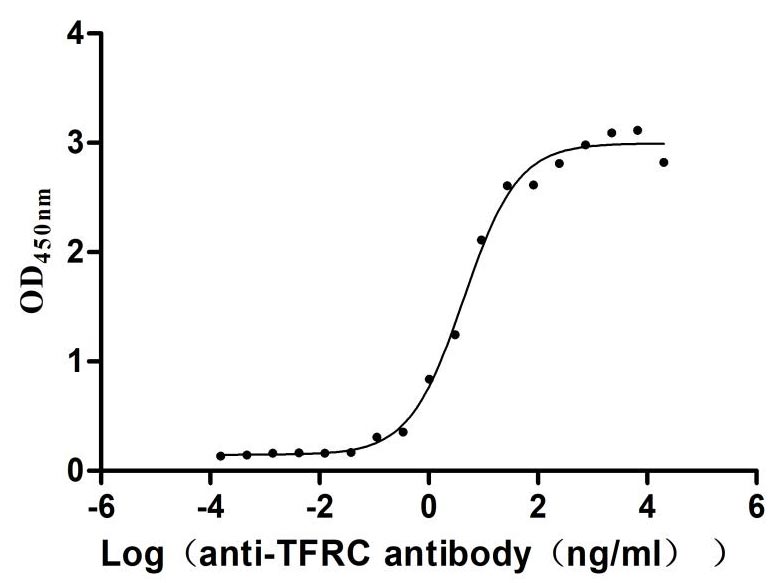
Recombinant Human Transferrin receptor protein 1 (TFRC), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
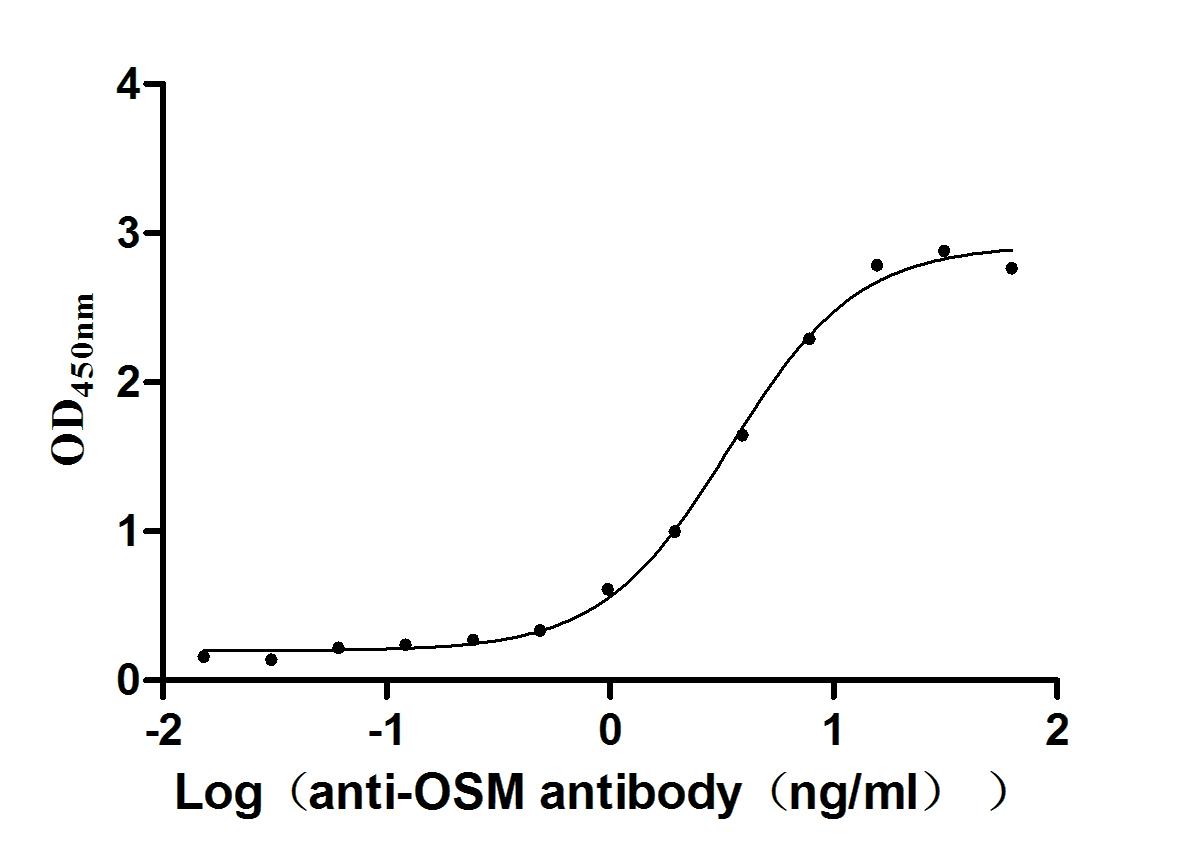
Recombinant Human Oncostatin-M (OSM), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
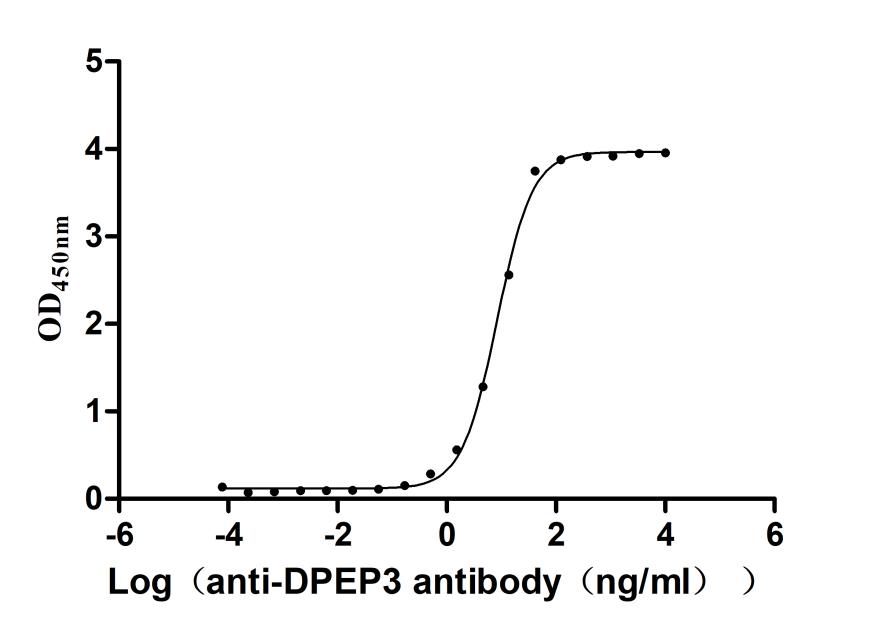
Recombinant Human Dipeptidase 3(DPEP3), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Macaca fascicularis Dipeptidase 3(DPEP3) (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
Recombinant DT3C (Diphtheria toxin & spg 3C domain) for Antibody Internalization Assay (Active)
Express system: E.coli
Species: N/A


-AC1.jpg)