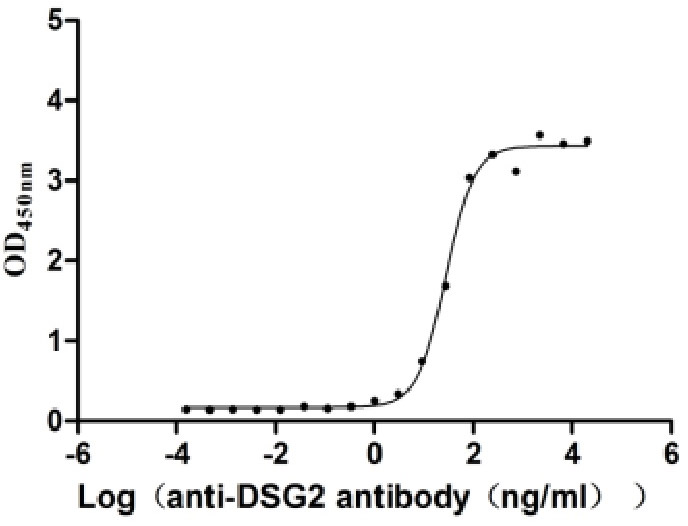
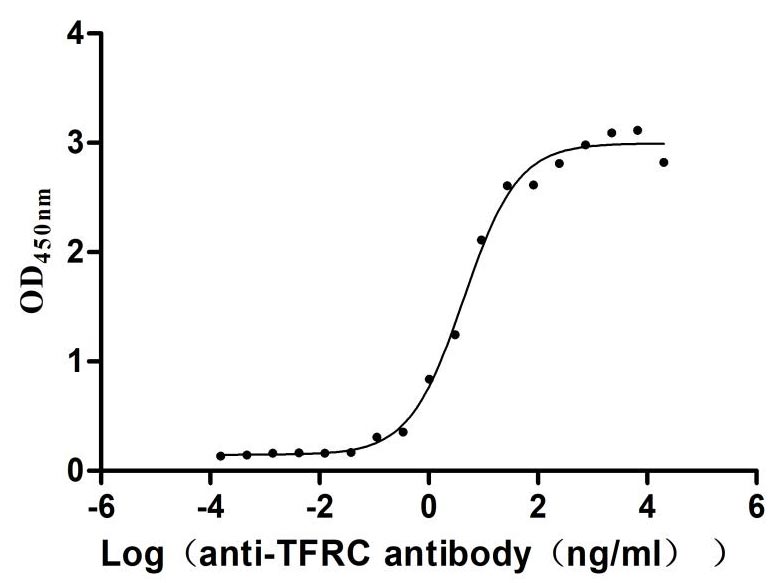
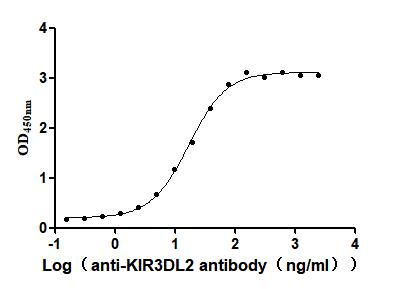
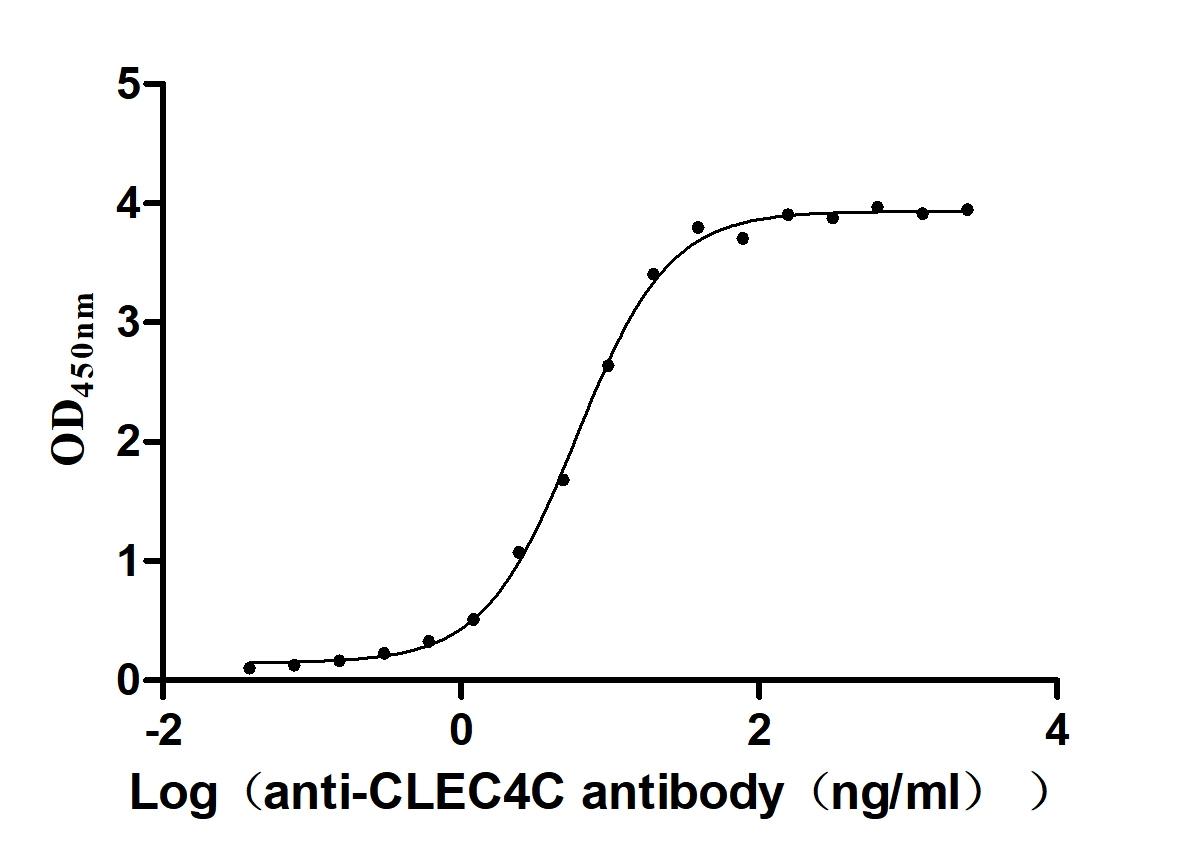
Recombinant Zika virus Genome polyprotein, partial
-
中文名称:
-
货号:CSB-EP3643GOZ3
-
规格:¥2328
-
图片:
-
其他:
产品详情
-
纯度:Greater than 85% as determined by SDS-PAGE.
-
基因名:N/A
-
Uniprot No.:
-
种属:Zika virus (ZIKV)
-
蛋白长度:Partial
-
来源:E.coli
-
分子量:28.2 kDa
-
表达区域:1415-1463aa+GGGGSGGGG+1499-1668aa
-
氨基酸序列KSVDMYIERAGDITWEKDAEVTGNSPRLDVALDESGDFSLVEEDGPPMRGGGGSGGGGSGALWDVPAPKEVKKGETTDGVYRVMTRRLLGSTQVGVGVMQEGVFHTMWHVTKGAALRSGEGRLDPYWGDVKQDLVSYCGPWKLDAAWDGLSEVQLLAVPPGERARNIQTLPGIFKTKDGDIGAVALDYPAGTSGSPILDKCGRVIGLYGNGVVIKNGSYVSAITQGKR
Note: The complete sequence including tag sequence, target protein sequence and linker sequence could be provided upon request. -
蛋白标签:N-terminal 6xHis-tagged
-
产品提供形式:Liquid or Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
缓冲液:If the delivery form is liquid, the default storage buffer is Tris/PBS-based buffer, 5%-50% glycerol. If the delivery form is lyophilized powder, the buffer before lyophilization is Tris/PBS-based buffer, 6% Trehalose.
-
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20°C/-80°C. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:13-23 business days
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet & COA:Please contact us to get it.
靶点详情
-
功能:Plays a role in virus budding by binding to the host cell membrane and packages the viral RNA into a nucleocapsid that forms the core of the mature virus particle. During virus entry, may induce genome penetration into the host cytoplasm after hemifusion induced by the surface proteins. Can migrate to the cell nucleus where it modulates host functions.; Inhibits RNA silencing by interfering with host Dicer.; Prevents premature fusion activity of envelope proteins in trans-Golgi by binding to envelope protein E at pH 6.0. After virion release in ...显示更多
-
基因功能参考文献:
- Here, by mutagenesis, the s found a major role of the N-glycosylation of flavivirus E protein in its transmission circle, facilitating its survival against the vector immune system during invasion of the mosquito midgut while blood feeding on the host. PMID: 29463651
- In complex with an inhibitor, the protease adopts a closed, "active" conformation with the NS2B chain wrapped around the NS3(pro) and contributing to the S2 pocket.[review] PMID: 29845530
- crystal structure of the apo ZIKV NS2B-NS3pro complex in a monomeric form; structure reveals a molecular mechanism for ZIKV NS3pro inhibition and identifies a new target for rational drug design against flavivirus PMID: 27752039
- Data indicate a crystal structure at 1.84 A resolution of ZIKV non-structural protein NS2B-NS3 protease with the last four amino acids of the NS2B cofactor bound at the NS3 active site. PMID: 27845325
- The potential roles of NS2B and NS4A Zika virus proteins in its global pandemic has been reported. PMID: 29428601
- The mutation enables NS1 binding to TBK1 and reduces TBK1 phosphorylation and inhibit interferon-beta induction. PMID: 29379028
- The Zika virus envelope protein glycan loop modulates antigenicity. PMID: 29304471
- Structural docking suggests that temoporfin potentially binds NS3 pockets that hold critical NS2B residues, thus inhibiting flaviviral polyprotein processing in a non-competitive manner. PMID: 28685770
- Sustained Specific and Cross-Reactive T Cell Responses to Zika and Dengue Virus NS3 in West Africa PMID: 29321308
- Based on the proposition that the Zika virus NS5 protein utilizes SIAH2-mediated proteasomal degradation of STAT2, an in-silico study was carried out to characterize the protein-protein interactions between NS5, SIAH2 and STAT2 proteins. PMID: 28365387
- As the NS2B co-factor is involved in substrate binding of flaviviral NS2B-NS3 proteases, the destabilization of the closed conformation in the linked construct makes it an attractive tool to search for inhibitors that interfere with the formation of the enzymatically active, closed conformation. PMID: 28336347
- Here, we solved the crystal structure of full-length NS1 protein, and found an extended membrane association interface contributed by the hydrophobic "spike" of a long intertwined loop, providing important information for ZIKV pathogenesis and development of diagnostic tools. PMID: 27578809
- When Zika virus NS5 was expressed, the formation of STAT1-STAT1 homodimers and their recruitment to IFN-gamma-stimulated genes, such as the gene encoding the proinflammatory cytokine CXCL10, were augmented. PMID: 28468880
- NS4A and NS4B, cooperatively suppress the Akt-mTOR pathway and lead to cellular dysregulation. PMID: 27524440
- The immature ZIKV contains a partially ordered capsid protein shell that is less prominent in other immature flaviviruses. PMID: 28067914
- The crystal structure of a C-terminal fragment of ZIKV nonstructural protein 1 (NS1), a major host-interaction molecule that functions in flaviviral replication, pathogenesis and immune evasion. PMID: 27088990
- the crystal structure of full-length Zika virus NS1 PMID: 27455458
- Study present the crystal structures of ZIKV NS5 N-terminal methyltransferase in complex with an RNA cap analogue ((m7)GpppA) and the free NS5 C-terminal RNA-dependent RNA polymerase. PMID: 28254839
- A high-resolution (1.62-A) crystal structure of the RNA helicase from the French Polynesia strain of Zika virus. PMID: 27399257
- The structure of ZIKV helicase-RNA has revealed that upon RNA binding, rotations of the motor domains can cause significant conformational changes. Strikingly, although ZIKV and dengue virus (DENV) apo-helicases share conserved residues for RNA binding, their different manners of motor domain rotations result in distinct individual modes for RNA recognition. PMID: 27430951
收起更多
-
亚细胞定位:[Capsid protein C]: Virion. Host nucleus. Host cytoplasm. Host cytoplasm, host perinuclear region.; [Peptide pr]: Secreted.; [Small envelope protein M]: Virion membrane; Multi-pass membrane protein. Host endoplasmic reticulum membrane; Multi-pass membrane protein.; [Envelope protein E]: Virion membrane; Multi-pass membrane protein. Host endoplasmic reticulum membrane; Multi-pass membrane protein.; [Non-structural protein 1]: Secreted. Host endoplasmic reticulum membrane; Peripheral membrane protein; Lumenal side.; [Non-structural protein 2A]: Host endoplasmic reticulum membrane; Multi-pass membrane protein.; [Serine protease subunit NS2B]: Host endoplasmic reticulum membrane; Multi-pass membrane protein.; [Serine protease NS3]: Host cytoplasm. Host endoplasmic reticulum membrane; Peripheral membrane protein; Cytoplasmic side.; [Non-structural protein 4A]: Host endoplasmic reticulum membrane; Multi-pass membrane protein.; [Non-structural protein 4B]: Host endoplasmic reticulum membrane; Multi-pass membrane protein.; [RNA-directed RNA polymerase NS5]: Host endoplasmic reticulum membrane; Peripheral membrane protein; Cytoplasmic side. Host nucleus.
-
数据库链接:
KEGG: vg:7751225



-AC1.jpg)