Recombinant Mouse NAD-dependent protein deacetylase sirtuin-7 (Sirt7)
-
货号:CSB-YP807372MO
-
规格:
-
来源:Yeast
-
其他:
-
货号:CSB-EP807372MO
-
规格:
-
来源:E.coli
-
其他:
-
货号:CSB-EP807372MO-B
-
规格:
-
来源:E.coli
-
共轭:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
货号:CSB-BP807372MO
-
规格:
-
来源:Baculovirus
-
其他:
-
货号:CSB-MP807372MO
-
规格:
-
来源:Mammalian cell
-
其他:
产品详情
-
纯度:>85% (SDS-PAGE)
-
基因名:Sirt7
-
Uniprot No.:
-
别名:Sirt7; Sir2l7; NAD-dependent protein deacetylase sirtuin-7; EC 2.3.1.286; NAD-dependent protein deacylase sirtuin-7; EC 2.3.1.-; Regulatory protein SIR2 homolog 7; SIR2-like protein 7
-
种属:Mus musculus (Mouse)
-
蛋白长度:full length protein
-
表达区域:1-402
-
氨基酸序列MAAGGGLSRS ERKAAERVRR LREEQQRERL RQVSRILRKA AAERSAEEGR LLAESEDLVT ELQGRSRRRE GLKRRQEEVC DDPEELRRKV RELAGAVRSA RHLVVYTGAG ISTAASIPDY RGPNGVWTLL QKGRPVSAAD LSEAEPTLTH MSITRLHEQK LVQHVVSQNC DGLHLRSGLP RTAISELHGN MYIEVCTSCI PNREYVRVFD VTERTALHRH LTGRTCHKCG TQLRDTIVHF GERGTLGQPL NWEAATEAAS KADTILCLGS SLKVLKKYPR LWCMTKPPSR RPKLYIVNLQ WTPKDDWAAL KLHGKCDDVM QLLMNELGLE IPVYNRWQDP IFSLATPLRA GEEGSHSRKS LCRSREEAPP GDQSDPLASA PPILGGWFGR GCAKRAKRKK VA
-
蛋白标签:Tag type will be determined during the manufacturing process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
产品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
靶点详情
-
功能:NAD-dependent protein-lysine deacylase that can act both as a deacetylase or deacylase (desuccinylase, depropionylase and deglutarylase), depending on the context. Specifically mediates deacetylation of histone H3 at 'Lys-18' (H3K18Ac). In contrast to other histone deacetylases, displays strong preference for a specific histone mark, H3K18Ac, directly linked to control of gene expression. H3K18Ac is mainly present around the transcription start site of genes and has been linked to activation of nuclear hormone receptors; SIRT7 thereby acts as a transcription repressor. Moreover, H3K18 hypoacetylation has been reported as a marker of malignancy in various cancers and seems to maintain the transformed phenotype of cancer cells. Also able to mediate deacetylation of histone H3 at 'Lys-36' (H3K36Ac) in the context of nucleosomes. Also mediates deacetylation of non-histone proteins, such as ATM, CDK9, DDX21, DDB1, FBL, FKBP5/FKBP51, GABPB1, RAN, RRP9/U3-55K and POLR1E/PAF53. Enriched in nucleolus where it stimulates transcription activity of the RNA polymerase I complex. Acts by mediating the deacetylation of the RNA polymerase I subunit POLR1E/PAF53, thereby promoting the association of RNA polymerase I with the rDNA promoter region and coding region. In response to metabolic stress, SIRT7 is released from nucleoli leading to hyperacetylation of POLR1E/PAF53 and decreased RNA polymerase I transcription. Required to restore the transcription of ribosomal RNA (rRNA) at the exit from mitosis. Promotes pre-ribosomal RNA (pre-rRNA) cleavage at the 5'-terminal processing site by mediating deacetylation of RRP9/U3-55K, a core subunit of the U3 snoRNP complex. Mediates 'Lys-37' deacetylation of Ran, thereby regulating the nuclear export of NF-kappa-B subunit RELA/p65. Acts as a regulator of DNA damage repair by mediating deacetylation of ATM during the late stages of DNA damage response, promoting ATM dephosphorylation and deactivation. May also deacetylate p53/TP53 and promotes cell survival, however such data need additional confirmation. Suppresses the activity of the DCX (DDB1-CUL4-X-box) E3 ubiquitin-protein ligase complexes by mediating deacetylation of DDB1, which prevents the interaction between DDB1 and CUL4 (CUL4A or CUL4B). Activates RNA polymerase II transcription by mediating deacetylation of CDK9, thereby promoting 'Ser-2' phosphorylation of the C-terminal domain (CTD) of RNA polymerase II. Deacetylates FBL, promoting histone-glutamine methyltransferase activity of FBL. Acts as a regulator of mitochondrial function by catalyzing deacetylation of GABPB1. Regulates Akt/AKT1 activity by mediating deacetylation of FKBP5/FKBP51. Required to prevent R-loop-associated DNA damage and transcription-associated genomic instability by mediating deacetylation and subsequent activation of DDX21, thereby overcoming R-loop-mediated stalling of RNA polymerases. In addition to protein deacetylase activity, also acts as protein-lysine deacylase. Acts as a protein depropionylase by mediating depropionylation of Osterix (SP7), thereby regulating bone formation by osteoblasts. Acts as a histone deglutarylase by mediating deglutarylation of histone H4 on 'Lys-91' (H4K91glu); a mark that destabilizes nucleosomes by promoting dissociation of the H2A-H2B dimers from nucleosomes. Acts as a histone desuccinylase: in response to DNA damage, recruited to DNA double-strand breaks (DSBs) and catalyzes desuccinylation of histone H3 on 'Lys-122' (H3K122succ), thereby promoting chromatin condensation and DSB repair. Also promotes DSB repair by promoting H3K18Ac deacetylation, regulating non-homologous end joining (NHEJ). Along with its role in DNA repair, required for chromosome synapsis during prophase I of female meiosis by catalyzing H3K18Ac deacetylation. Involved in transcriptional repression of LINE-1 retrotransposon via H3K18Ac deacetylation, and promotes their association with the nuclear lamina. Required to stabilize ribosomal DNA (rDNA) heterochromatin and prevent cellular senescence induced by rDNA instability. Acts as a negative regulator of SIRT1 by preventing autodeacetylation of SIRT1, restricting SIRT1 deacetylase activity.
-
基因功能参考文献:
- The results indicate that miR-148b-3p contributes to the regulation of hypoxia/reoxygenation-induced cardiomyocyte apoptosis in vitro through targeting SIRT7 and modulating p53-mediated pro-apoptotic signaling. PMID: 30308185
- Data indicate that Sirt7 has a slight protective impact on both the adaptive immune system and neurogenesis. However, overall this epigenetic factor is not capable of impacting the acute or chronic phase of neuroinflammation. PMID: 28939414
- Loss of SIRT7 is associated with Impairment of Adipogenesis and increased Sirt1 Activity. PMID: 28923965
- the results of our study suggest that SIRT7 is involved in protecting neurons against OGD/R-induced injury, possibly through regulation of the p53-mediated proapoptotic signaling pathway, indicating a potential therapeutic target for cerebral ischemia/reperfusion injury. PMID: 28675767
- Histological analysis revealed that there is no apparent structural abnormality of the amygdala and hippocampus, which are regions involved in fear memory consolidation, in Sirt7 KO mice. Our results indicate that SIRT7 is involved in the consolidation of fear memory. PMID: 29101029
- the decline in SIRT7 in lung fibroblasts has a profibrotic effect, which is mediated by changes in Smad3 levels. PMID: 28385812
- SIRT7 inhibits TR4 degradation by deacetylation of DDB1. PMID: 28623141
- These results reveal a direct role for SIRT7 in double-strand break repair and establish a functional link between SIRT7-mediated histone H3 K18 deacetylation and the maintenance of genome integrity. PMID: 27225932
- miR-93 inhibits the metabolic target Sirt7, which we identified as a major driver of in vivo adipogenesis via induction of differentiation and maturation of early adipocyte precursors. PMID: 26321631
- The role of SIRT7 has emerged in protein synthesis, chromatin remodelling, cellular survival and lipid metabolism. PMID: 25435428
- SIRT7 stabilizes and activates the GABP alpha/GABPbeta complex via deacetylation of GABPbeta1. PMID: 25200183
- SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. PMID: 24703702
- Study identifies SIRT7 as a cofactor of Myc for transcriptional repression and delineates a druggable regulatory branch of the ER stress response that prevents and reverts fatty liver disease. PMID: 24210820
- Mybbp1a is a novel negative regulator of Sirt7. PMID: 24134843
- SirT7 plays a dominant role in the growth inhibition of the P19 cells. PMID: 20078942
- SIRT7 is a positive regulator of Pol I transcription and is required for cell viability in mammals. PMID: 16618798
- Suggest a critical role of Sirt7 in the regulation of stress responses and cell death in the heart. Enhanced p53 activation by lack of Sirt7-mediated deacetylation contributes to the heart phenotype of Sirt7 mutant mice. PMID: 18239138
- Using Sirt7 knockout and overexpressing cells we demonstrate an anti-proliferative role of Sirt7. We also show that Sirt7 expression inversely correlates with the tumorigenic potential of several murine cell lines PMID: 19261981
显示更多
收起更多
-
亚细胞定位:Nucleus, nucleolus. Nucleus, nucleoplasm. Chromosome. Cytoplasm.
-
蛋白家族:Sirtuin family, Class IV subfamily
-
组织特异性:Detected in liver, spleen and testis. Detected in embryos.
-
数据库链接:
KEGG: mmu:209011
STRING: 10090.ENSMUSP00000079093
UniGene: Mm.292957
Most popular with customers
-
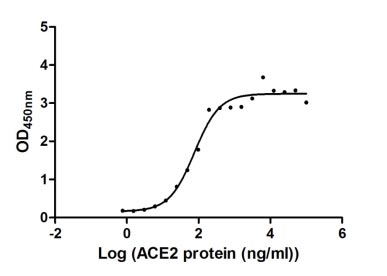
Recombinant Human Angiotensin-converting enzyme 2 (ACE2), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Tumor necrosis factor ligand superfamily member 14 (TNFSF14), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Tissue factor pathway inhibitor (TFPI), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Claudin-6 (CLDN6)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Microtubule-associated protein tau (MAPT) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
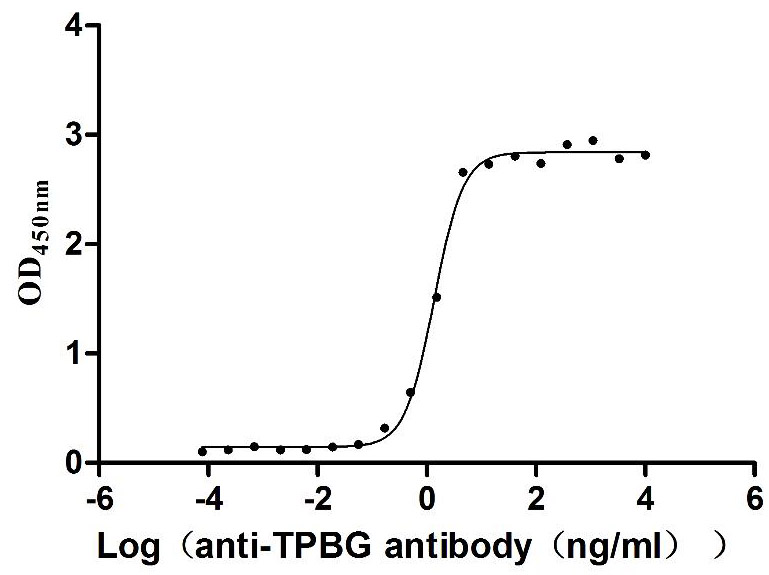
Recombinant Human Trophoblast glycoprotein (TPBG), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
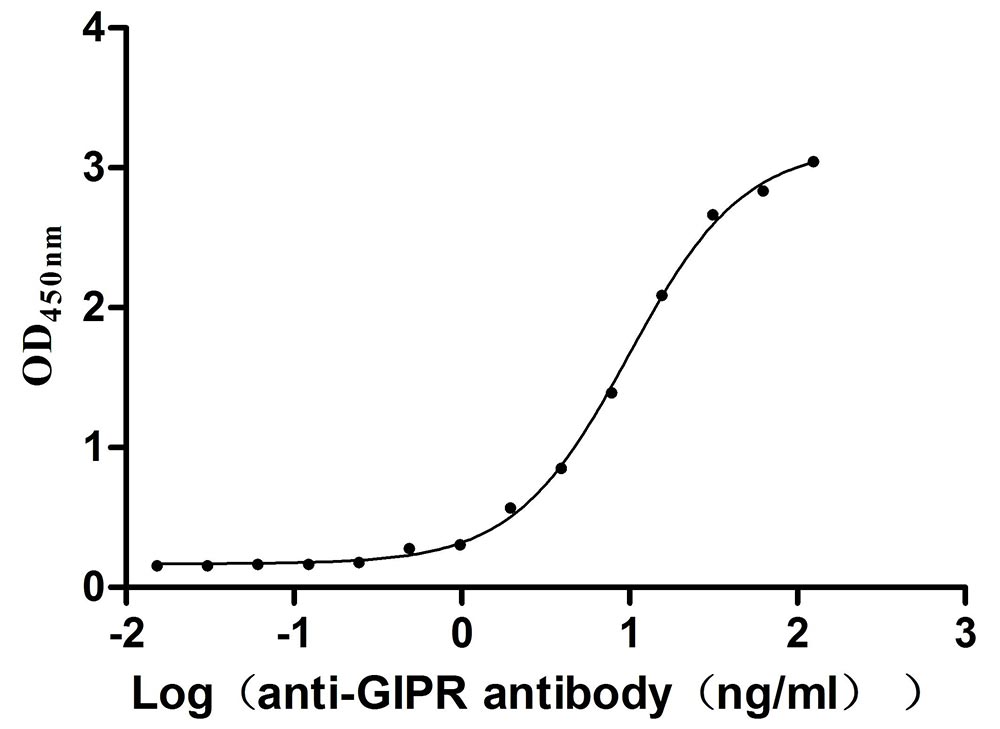
Recombinant Mouse Gastric inhibitory polypeptide receptor (Gipr), partial (Active)
Express system: Mammalian cell
Species: Mus musculus (Mouse)
-
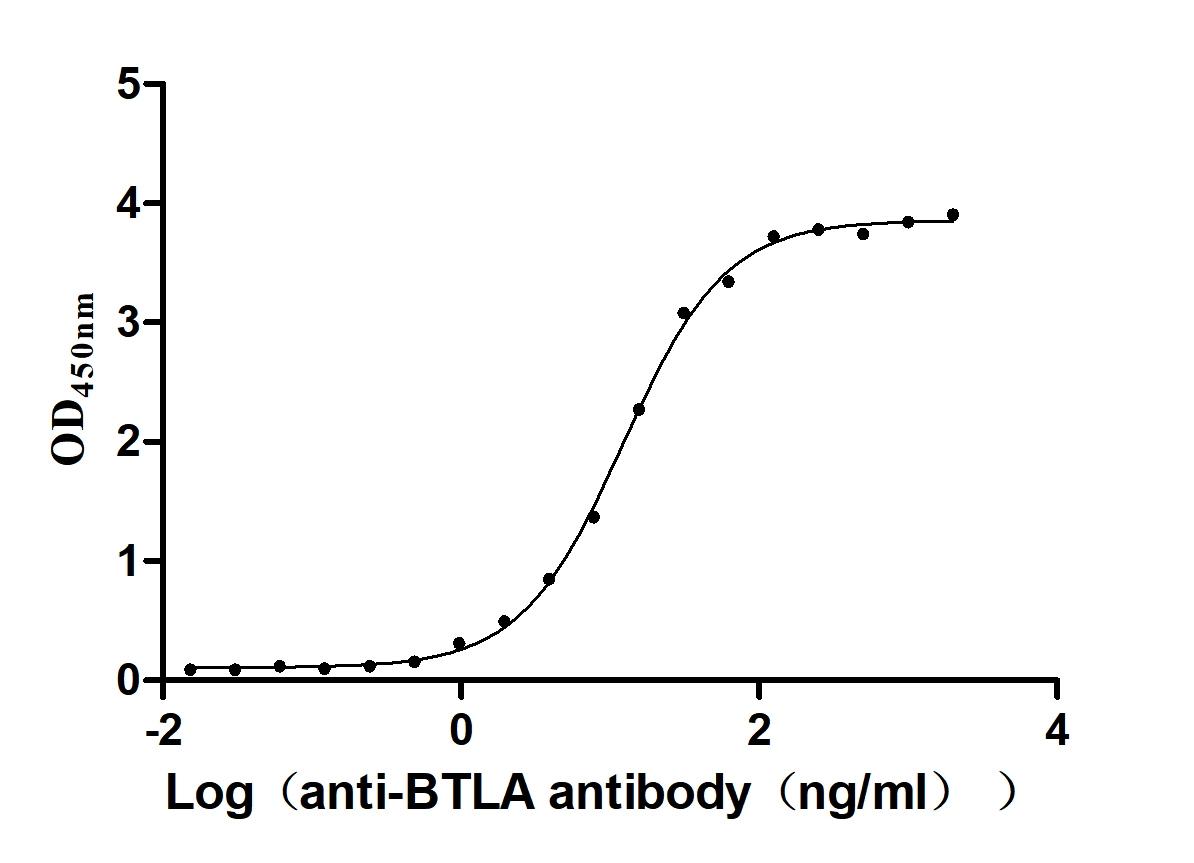
Recombinant Human B- and T-lymphocyte attenuator(BTLA), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)


-AC1.jpg)
-AC1.jpg)