Recombinant Human Crossover junction endonuclease MUS81 (MUS81)
-
中文名称:人MUS81重组蛋白
-
货号:CSB-YP836272HU
-
规格:
-
来源:Yeast
-
其他:
-
中文名称:人MUS81重组蛋白
-
货号:CSB-EP836272HU
-
规格:
-
来源:E.coli
-
其他:
-
中文名称:人MUS81重组蛋白
-
货号:CSB-EP836272HU-B
-
规格:
-
来源:E.coli
-
共轭:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
中文名称:人MUS81重组蛋白
-
货号:CSB-BP836272HU
-
规格:
-
来源:Baculovirus
-
其他:
-
中文名称:人MUS81重组蛋白
-
货号:CSB-MP836272HU
-
规格:
-
来源:Mammalian cell
-
其他:
产品详情
-
纯度:>85% (SDS-PAGE)
-
基因名:MUS81
-
Uniprot No.:
-
别名:Crossover junction endonuclease MUS81; FLJ21012; FLJ44872; Mus 81; MUS81; MUS81 Endonuclease Homolog ; MUS81_HUMAN
-
种属:Homo sapiens (Human)
-
蛋白长度:full length protein
-
表达区域:1-551
-
氨基酸序列MAAPVRLGRK RPLPACPNPL FVRWLTEWRD EATRSRRRTR FVFQKALRSL RRYPLPLRSG KEAKILQHFG DGLCRMLDER LQRHRTSGGD HAPDSPSGEN SPAPQGRLAE VQDSSMPVPA QPKAGGSGSY WPARHSGARV ILLVLYREHL NPNGHHFLTK EELLQRCAQK SPRVAPGSAR PWPALRSLLH RNLVLRTHQP ARYSLTPEGL ELAQKLAESE GLSLLNVGIG PKEPPGEETA VPGAASAELA SEAGVQQQPL ELRPGEYRVL LCVDIGETRG GGHRPELLRE LQRLHVTHTV RKLHVGDFVW VAQETNPRDP ANPGELVLDH IVERKRLDDL CSSIIDGRFR EQKFRLKRCG LERRVYLVEE HGSVHNLSLP ESTLLQAVTN TQVIDGFFVK RTADIKESAA YLALLTRGLQ RLYQGHTLRS RPWGTPGNPE SGAMTSPNPL CSLLTFSDFN AGAIKNKAQS VREVFARQLM QVRGVSGEKA AALVDRYSTP ASLLAAYDAC ATPKEQETLL STIKCGRLQR NLGPALSRTL SQLYCSYGPL T
-
蛋白标签:Tag type will be determined during the manufacturing process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
产品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
相关产品
靶点详情
-
功能:Interacts with EME1 and EME2 to form a DNA structure-specific endonuclease with substrate preference for branched DNA structures with a 5'-end at the branch nick. Typical substrates include 3'-flap structures, replication forks and nicked Holliday junctions. May be required in mitosis for the processing of stalled or collapsed replication forks.
-
基因功能参考文献:
- Replication fork progression in BRCA2-deficient cells requires MUS81. MUS81 nucleolytic activity is required to activate compensatory DNA synthesis during mitosis and to resolve mitotic interlinks, thus facilitating chromosome segregation. PMID: 28714477
- Using RNAi or FA-P cells complemented with SLX4 mutants that abrogate interaction with MUS81 or SLX1, we show that SLX4 cooperates with MUS81 to introduce DSBs after replication stress but also counteracts pathological targeting of demised forks by GEN1. PMID: 28290553
- The results showed that MUS81 modulates MCM2 levels as well as homologous recombination (HR) activity. Moreover, downregulation of MUS81 increased the sensitivity of epithelial ovarian cancer (EOC)cells to olaparib by inducing S phase arrest and promoting apoptosis through activation of MCM2. MUS81 may be a potential novel therapeutic target for EOC. PMID: 29393493
- Mus81 knockdown improves the chemosensitivity of colon cancer cells by inducing S phase arrest and promoting apoptosis through activating CHK1 pathway PMID: 28291626
- Low EZH2 or MUS81 expression levels predict chemoresistance. PMID: 29035360
- RECQ5 removes RAD51 filaments stabilizing stalled replication forks at common fragile sites and hence facilitates CFS cleavage by MUS81-EME1. PMID: 28575661
- The mitotic DNA synthesis is RAD52 dependent, and RAD52 is required for the timely recruitment of MUS81 and POLD3 to common fragile sites in early mitosis. PMID: 27984745
- Data suggest that dimeric GEN1 binds with high affinity/selectivity to Holliday junctions, introducing two symmetrical hydrolytic cleavages of phosphodiester backbone; at present, less is known about SLX1-SLX4-MUS81-EME1 resolving enzyme complex. (GEN1 = Holliday junction 5' flap endonuclease; SLX = structure-specific endonuclease subunit; MUS81 = MUS81 endonuclease; EME1 = essential meiotic endonuclease 1) [REVIEW] PMID: 27990631
- down-regulation of MUS81 expression in ovarian cancer cells inhibited cell proliferation and colony formation ability, and influenced cell cycle progression PMID: 27255997
- SLX4-SLX1 Protein-independent Down-regulation of MUS81-EME1 Protein by HIV-1 Viral Protein R (Vpr). PMID: 27354282
- Mus81 sumoylation is important for normal mitotic chromosome congression. PMID: 28318385
- Data suggest that the ATM/Chk2 may promote the repair of DNA damage caused by cisplatin by sustaining methyl methanesulfonate and ultraviolet-sensitive gene clone 81, and the double-strand breaks generated by methyl methanesulfonate and ultraviolet-sensitive gene clone 81 may activate the ATM/Chk2 pathway. PMID: 28347251
- Avoiding damage formation through invalidation of Mus81-Eme2 and Mre11, or preventing damage signaling by turning off the ATM pathway, suppresses the replication phenotypes of Chk1-deficient cells. PMID: 26804904
- Mus81 knockdown improves the chemosensitivity of HCC cells by inducing S-phase arrest and promoting apoptosis through CHK1 pathway, suggesting Mus81 as a novel therapeutic target for HCC. PMID: 26714930
- Mus81 regulates the rate of DNA replication during normal growth by promoting replication fork progression while reducing the frequency of replication initiation events. PMID: 25879486
- Identification and characterization of MUS81 point mutations that abolish interaction with the SLX4 scaffold protein.MUS81 function in DNA interstrand crosslinks repair requires interaction with SLX4. PMID: 25224045
- The data highlight the importance of Mus81 and Blm in DNA double-strand repair pathways, fertility, development and cancer. PMID: 24858046
- s confirmed that HIV-1 Vpr induces degradation of Mus81 although, surprisingly, degradation is independent and genetically separable from Vprs ability to induce G2 arrest. PMID: 25618414
- Results define distinct and temporal roles for MUS81-EME1 and MUS81-EME2 in the maintenance of genome stability. PMID: 24813886
- Our findings reveal a novel RAD52/MUS81-dependent mechanism that promotes cell viability and genome integrity in checkpoint-deficient cells, and disclose the involvement of MUS81 to multiple processes after replication stress PMID: 24204313
- While Mus81-Eme1 shares several common features with members of the 5' flap nuclease family, the combined structural, biochemical, and biophysical analyses explain why Mus81-Eme1 preferentially cleaves 3' flap DNA substrates with 5' nicked ends. PMID: 24733841
- In vivo HJ resolution depends on both SLX4-associated MUS81-EME1 and SLX1, suggesting that they are acting in concert in the context of SLX4. PMID: 24080495
- The presence of a 5' phosphate terminus at nicks and gaps rendered DNA significantly less susceptible to the cleavage by MUS81-EME2 than its absence. PMID: 24692662
- This study identified a winged helix domain within the N-terminal region of human MUS81 that binds DNA, increases the activity of MUS81-EME1/EME2 complexes and influences the incision position of MUS81-EME2 but not MUS81-EME1 complexes on synthetic forks, 3' flaps and nicked Holliday junctions. PMID: 23982516
- Data show that three structure-selective endonucleases, SLX1-SLX4, MUS81-EME1, and GEN1, define two pathways of Holliday junctions (HJs) resolution in HeLa cells. PMID: 24076221
- the DNA structure-specific nuclease MUS81-EME1 localizes to CFS loci in early mitotic cells, and promotes the cytological appearance of characteristic gaps or breaks observed at CFSs in metaphase chromosomes. PMID: 23811685
- FBH1 helicase activity is required to eliminate cells with excessive replication stress through the generation of MUS81-induced DNA double-strand breaks. PMID: 23361013
- we demonstrate that double-strand breaks, observed upon oncogene over-expression, depend on the MUS81 endonuclease, which represents a parallel pathway collaborating with WRN to prevent cell death. PMID: 22410776
- Data show that Mus81/Eme1-dependent DNA damage--rather than a global increase in replication-fork stalling--is the cause of incomplete replication in Chk1-deficient cells. PMID: 21858151
- Mus81 cleaves stalled replication forks, which allows dissipation of the excessive supercoiling resulting from Top1 inhibition, spontaneous reversal of Top1cc, and replication fork progression. PMID: 22123861
- Results demonstrate a novel role of Wee1 in controlling Mus81-Eme1 and DNA replication in human cells. PMID: 21859861
- Mus81 is a novel prognostic marker for colorectal carcinoma PMID: 21175991
- Mus81 down-regulation correlated significantly to invasion depth (p = 0.015) and poorly-differentiated type (p = 0.016) of gastric cancer. PMID: 21187482
- Mus81-associated endonuclease may play a more direct role in replication fork collapse by catalysing the cleavage of stalled fork structures. PMID: 12374758
- hMUS81.hMMS4 complex is a structure-specific nuclease that is capable of resolving fork structure [hMMS4] PMID: 12686547
- Mus81 binds to a homolog of fission yeast Eme1 in vitro and in vivo and Mus81-Eme1 resolves Holliday junctions in vivo. PMID: 14617801
- Mus81 is required to repair problems that arise most frequently in the highly repetitive nucleolar DNA. PMID: 14638871
- Data suggest a new function of BLM in cooperating with Mus81 during processing and restoration of stalled replication forks. PMID: 15805243
- Mus81 deficiency affects cell cycle progression and promotes DNA rereplication. PMID: 16456034
- Pulsed-field gel electrophoresis of the mus81Delta mutant revealed an expansion of the rDNA locus depending on RAD52, in addition to fragmentation of Chr XII in the sgs1Deltamus81(ts) mutant at permissive temperature. PMID: 17555773
- Mus81 and FANCB have different roles in repair of DNA damage during replication in human cells PMID: 17903171
- Data suggest that Mus81 suppresses chromosomal instability by converting potentially detrimental replication-associated DNA structures into intermediates that are more amenable to DNA repair. PMID: 17934473
- BLM helicase and Mus81 are required to induce transient double-stranded DNA breaks in response to DNA replication stress. PMID: 18054789
- Mus81-Eme1 can ensure coordinate, bilateral cleavage of Holliday junction-like structures. PMID: 18310322
- the crystal structure of the Mus81-Eme1 complex PMID: 18413719
- Mutations and abnormal expression of MUS81 gene in the LSCC tissues were observed. PMID: 18841572
- WRN is required to avoid accumulation of double-strand breaks and fork collapse after replication perturbation, and that prompt MUS81-dependent generation of DSBs is instrumental for recovery from hydroxyurea-mediated replication arrest. PMID: 18852298
- results demonstrate a link between branch migration activity of hRad54 and structure-specific endonuclease activity of hMus81-Eme1, suggesting that the Rad54 and Mus81-Eme1 proteins may cooperate in the processing of Holliday junction-like intermediates PMID: 19017809
- MUS81 is involved in the maintenance of ALT (alternative lengthening of telomeres) cell survival at least in part by homologous recombination of telomeres. PMID: 19363487
显示更多
收起更多
-
亚细胞定位:Nucleus, nucleolus. Note=Recruited to foci of DNA damage in S-phase cells.
-
蛋白家族:XPF family
-
组织特异性:Widely expressed.
-
数据库链接:
HGNC: 29814
OMIM: 606591
KEGG: hsa:80198
STRING: 9606.ENSP00000307853
UniGene: Hs.288798
Most popular with customers
-
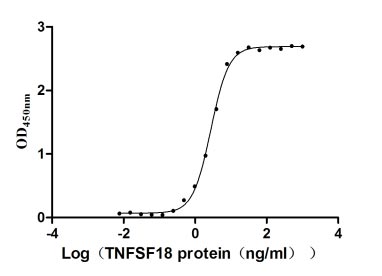
Recombinant Human Tumor necrosis factor ligand superfamily member 18 (TNFSF18), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
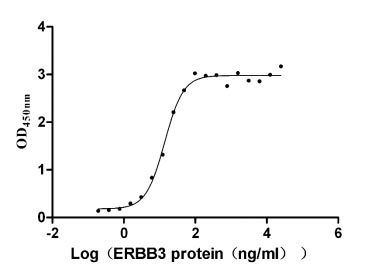
Recombinant Human Receptor tyrosine-protein kinase erbB-3 (ERBB3), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
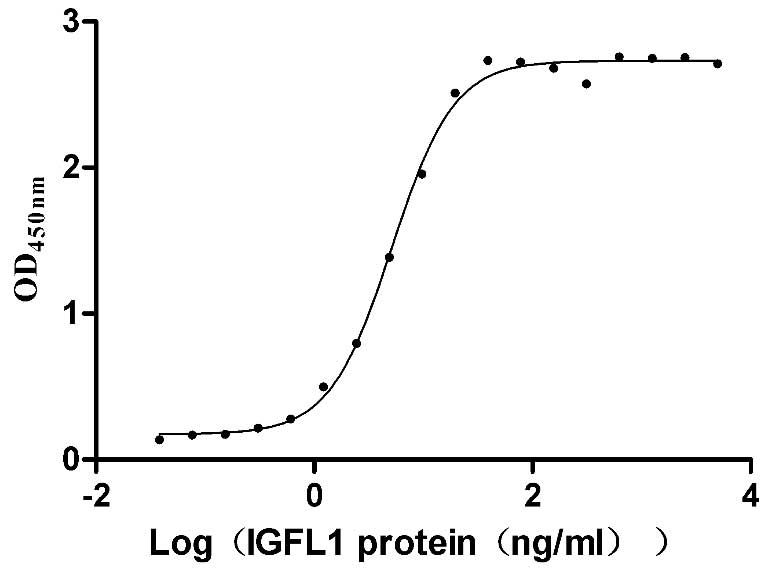
Recombinant Human IGF-like family receptor 1 (IGFLR1), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
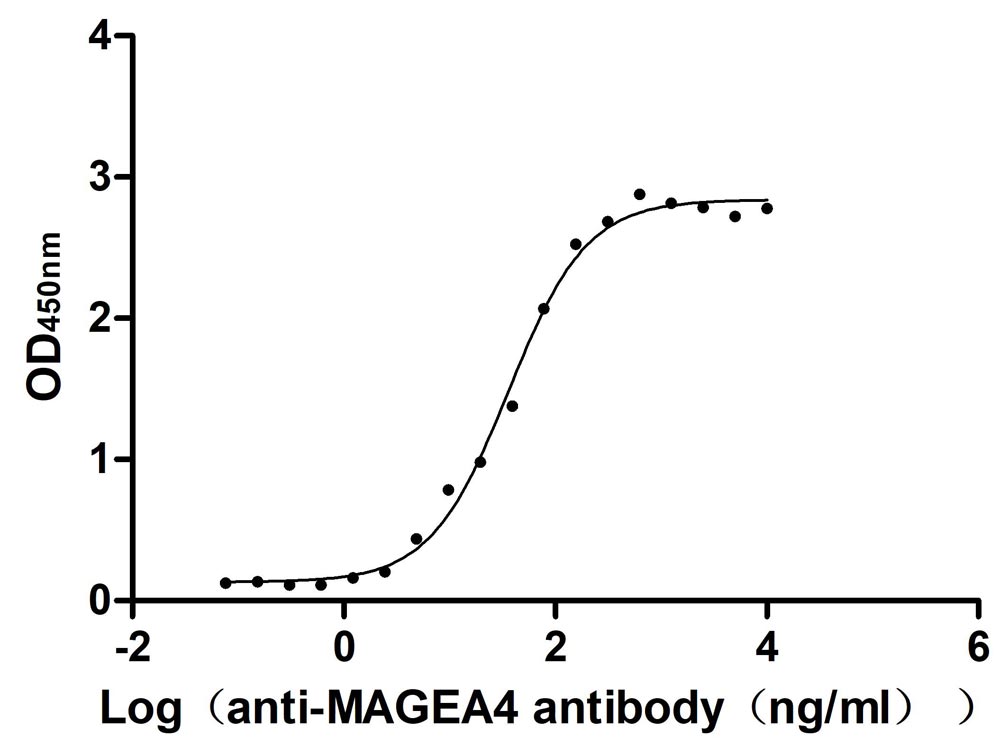
Recombinant Human Melanoma-associated antigen 4 (MAGEA4) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Intestinal-type alkaline phosphatase (ALPI) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
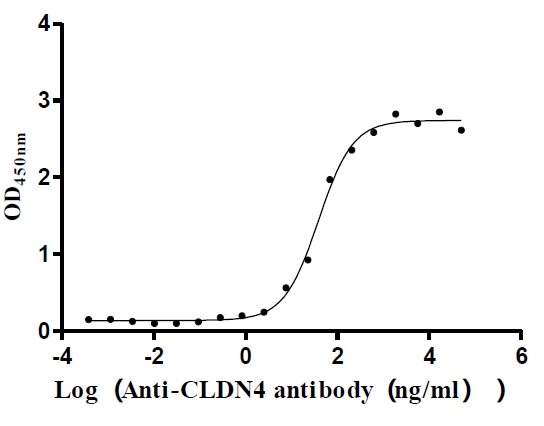
Recombinant Human Claudin-4 (CLDN4)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Claudin-6 (CLDN6)-VLPs, Fluorescent (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
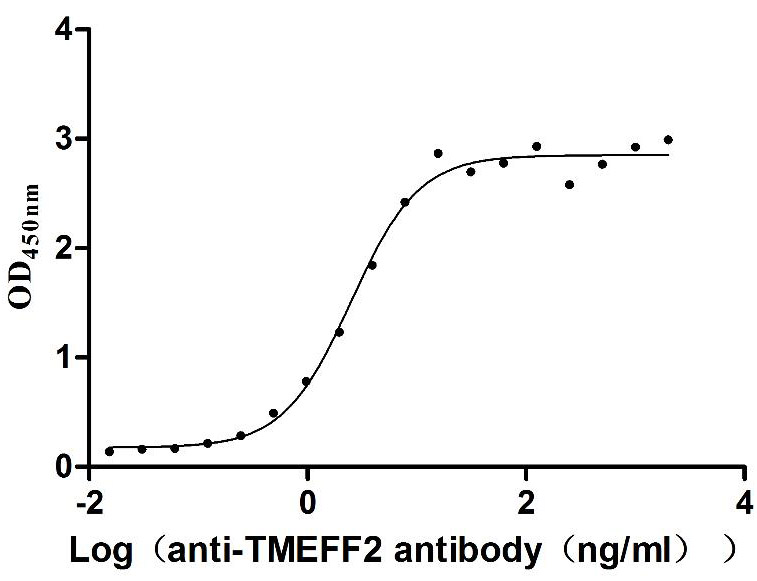
Recombinant Human Tomoregulin-2 (TMEFF2), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)



f4-AC1.jpg)