Recombinant Escherichia coli Holliday junction ATP-dependent DNA helicase RuvA (ruvA)
In Stock-
中文名称:大肠杆菌ruvA重组蛋白
-
货号:CSB-EP359062ENV
-
规格:¥2328
-
图片:
-
其他:
产品详情
-
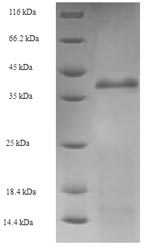
纯度:Greater than 90% as determined by SDS-PAGE.
-
基因名:ruvA
-
Uniprot No.:
-
别名:ruvA; b1861; JW1850; Holliday junction ATP-dependent DNA helicase RuvA; EC 3.6.4.12
-
种属:Escherichia coli (strain K12)
-
蛋白长度:Full Length
-
来源:E.coli
-
分子量:38.1kDa
-
表达区域:1-203aa
-
氨基酸序列MIGRLRGIIIEKQPPLVLIEVGGVGYEVHMPMTCFYELPEAGQEAIVFTHFVVREDAQLLYGFNNKQERTLFKELIKTNGVGPKLALAILSGMSAQQFVNAVEREEVGALVKLPGIGKKTAERLIVEMKDRFKGLHGDLFTPAADLVLTSPASPATDDAEQEAVAALVALGYKPQEASRMVSKIARPDASSETLIREALRAAL
Note: The complete sequence including tag sequence, target protein sequence and linker sequence could be provided upon request. -
蛋白标签:N-terminal 6xHis-SUMO-tagged
-
产品提供形式:Liquid or Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
缓冲液:Tris-based buffer,50% glycerol
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:3-7 business days
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet & COA:Please contact us to get it.
相关产品
靶点详情
-
功能:The RuvA-RuvB complex in the presence of ATP renatures cruciform structure in supercoiled DNA with palindromic sequence, indicating that it may promote strand exchange reactions in homologous recombination. RuvAB is a helicase that mediates the Holliday junction migration by localized denaturation and reannealing. RuvA stimulates, in the presence of DNA, the weak ATPase activity of RuvB. Binds both single- and double-stranded DNA (dsDNA). Binds preferentially to supercoiled rather than to relaxed dsDNA.
-
基因功能参考文献:
- Regression of replication forks stalled by leading-strand template damage: both RecG and RuvAB catalyze regression, but RuvC cleaves the holliday junctions formed by RecG preferentially. PMID: 25138216
- These results suggest that RecQ acts upstream of RuvABC, RecG and XerC proteins, a finding that is compatible with its primary role in initiation of the RecF recombination pathway. PMID: 24036154
- RNA polymerase mutations reduce the synergism between priB and ruvABC. PMID: 22957744
- DNA fragmentation absolutely depends on both RecA-catalyzed homologous strand exchange and RuvABC-catalyzed Holliday junction resolution PMID: 22194615
- Role in converting the initial single-strand DNA-protein cleavage complex into a double-strand break prior to repair by homologous recombination. PMID: 20601468
- Study hypothesized that RuvAB catalyzes replication fork reversal in order to produce SOS(Con) expression. PMID: 20304994
- RuvA octamerization is essential for the full biological activity of RuvABC. PMID: 15556943
- RuvA is less mobile at a heterologous junction compared to a homologous junction, as two opposing DnaB pumps are required to mobilize RuvA over heterologous DNA. PMID: 16324713
- RuvABC is required to resolve junctions that arise during the repair of a subset of nonarresting lesions after replication has passed through the template PMID: 16895921
- The s present here the isolation and characterization of ruvA and ruvB single mutants that are impaired for replication fork reversal at forks arrested by the inactivation of polymerase III, while they remain capable of homologous recombination. PMID: 18942176
显示更多
收起更多
-
亚细胞定位:Cytoplasm. Note=In 15% of cell localizes to discrete nucleoid foci (probable DNA damage sites) upon treatment with mitomycin C (MMC) for 2 hours.
-
蛋白家族:RuvA family
-
数据库链接:
KEGG: ecj:JW1850
STRING: 316385.ECDH10B_2002
Most popular with customers
-
Recombinant Human Tumor necrosis factor ligand superfamily member 9 (TNFSF9), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Mouse Transthyretin (Ttr) (Active)
Express system: Mammalian cell
Species: Mus musculus (Mouse)
-
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
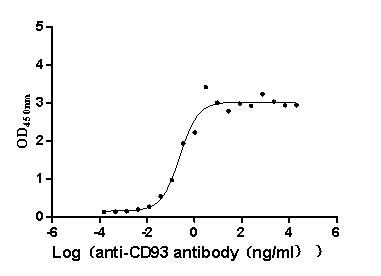
Recombinant Macaca fascicularis CD93 molecule (CD93), partial (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
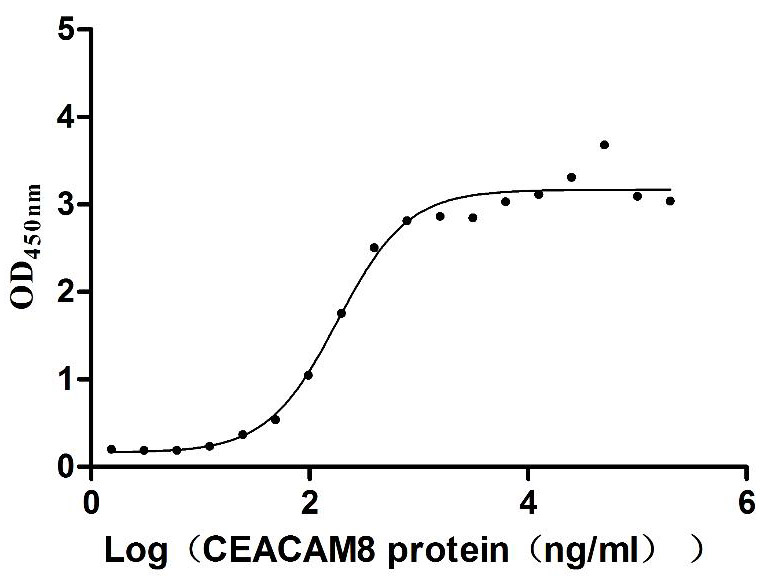
Recombinant Human Carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
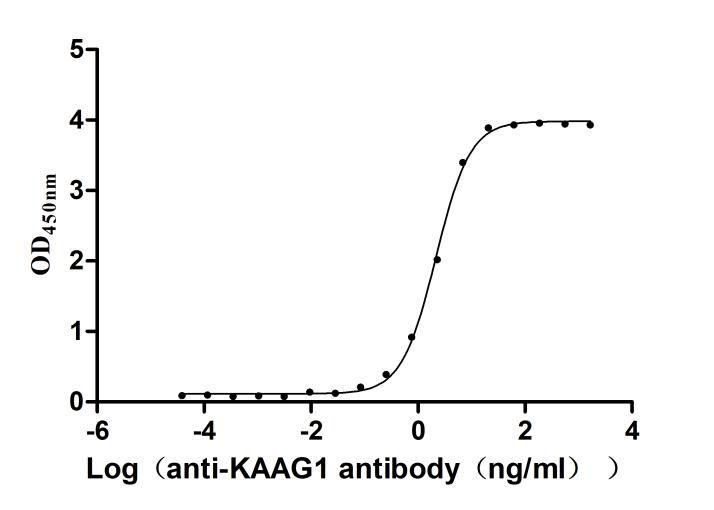
Recombinant Human Kidney-associated antigen 1(KAAG1) (Active)
Express system: E.coli
Species: Homo sapiens (Human)
-
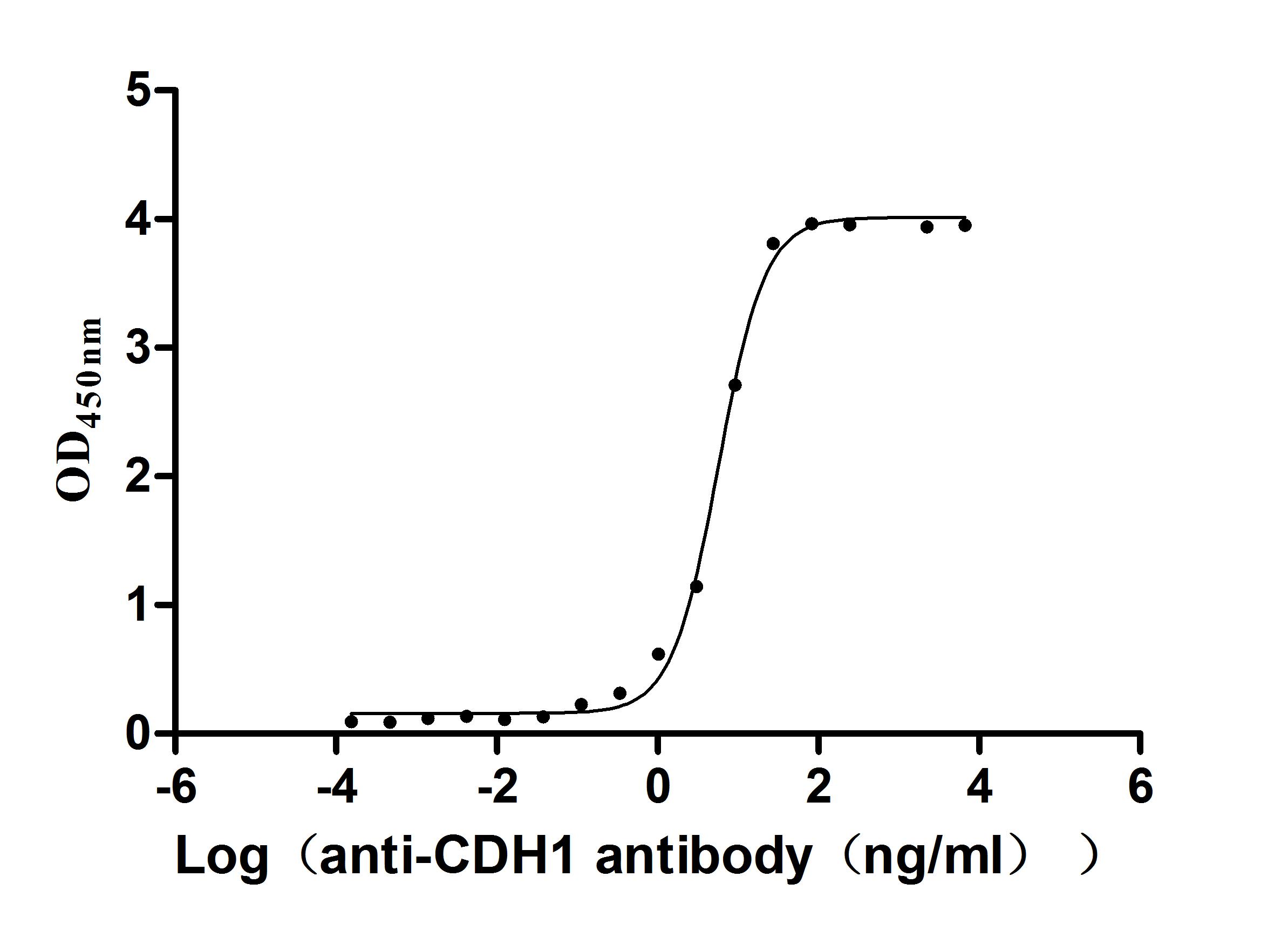
Recombinant Human Cadherin-1(CDH1),partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Mouse Cadherin-6(Cdh6),partial (Active)
Express system: Mammalian cell
Species: Mus musculus (Mouse)